Research Bio
Andy Martin is a Howard Hughes Medical Institute Investigator and Professor of Biochemistry, Biophysics and Structural Biology. His research interests:
In all cells, the degradation of intracellular proteins is highly specific and tightly regulated by ATP-dependent compartmental proteases. These proteases belong to the AAA+ ATPase family, whose characteristic feature is a structurally conserved ATPase domain that assembles into oligomeric rings and converts the energy of ATP binding and hydrolysis into conformational changes to perform mechanical work. Molecular machines of this enzyme family are also active, for instance, as protein translocases, clamp loaders, helicases, and mitochondrial motor proteins. For protein degradation, the AAA+ ATPases collaborate with compartmental peptidases. In these complexes, the ATPases recognize appropriate protein substrates and harness ATP hydrolysis to drive the mechanical unfolding and translocation of the polypeptide chain into a sequestered degradation chamber for proteolytic cleavage.
The research of the Martin lab focuses to a large extent on the 26S proteasome, the major ATP-dependent protease in eukaryotic cells. In addition to processing misfolded, damaged, and incompletely translated proteins, the proteasome selectively degrades a large variety of regulatory proteins that are involved in cell-cycle control, cell differentiation, signal transduction, transcription, and other vital processes, making it a very important mediator of post-translational control.
Most proteasomal substrates are marked for degradation by the attachment of a poly-ubiquitin chain, which acts as a tethering signal for substrate delivery. Although substantial knowledge about the ubiquitin-tagging systems has already been accumulated, the mechanisms underlying proteasomal substrate degradation, the irreversible and hence critical step for the unidirectional control of cellular pathways, remain largely elusive. In addition, their are several other AAA+ motor proteins, including p97/Cdc48 or Msp1, that act upstream of the proteasome to facilitate the quality control and degradation of damaged, mislocalized, or obsolete proteins.
The goals of the Martin are to decipher the fundamental principles that govern substrate engagement, de-ubiquitylation, unfolding, and translocation by the proteasome, the modulation of these activities by various adaptors and cofactors, the regulation of the proteasome, for instance by the "ubiquitin code", and the mechanism of related AAA+ motor proteins. The lab uses various approaches biochemistry, biophysics, and structural biology, including cryo-electron microscopy, X-ray crystallography, fluorescence and FRET-based measurements both in bulk and at the single-molecule level through TIRF microscopy, dual trap optical tweezing, hydrogen-deuterium exchange combined with mass spectrometry, and in vivo crosslinking / mass spec. The Martin lab also uses its biochemical tools and robust biophysical assays for high-throughput screenings to develop novel inhibitors of the 26S proteasome or lead compounds that can be further developed as PROTACs or molecular glues for targeted protein degradation.
Research Expertise and Interest
26S proteasome, Ubiquitin-Proteasome System, AAA+ ATPases, Molecular motor proteins
In the News
Glaunsinger, Martin named Howard Hughes Medical Institute investigators
Andreas Martin, an associate professor of molecular and cell biology, and Britt Glaunsinger, an associate professor of plant and microbial biology, are the campus’s newest Howard Hughes Medical Institute investigators.
Cancer’s Disposal System: Target for a Cure?
Andreas Martin has developed novel systems and strategies to screen for compounds that selectively inhibit protein turnover in the cell and may lead to new drugs against cancer.
New Information on the Waste-Disposal Units of Living Cells
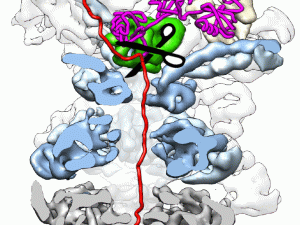
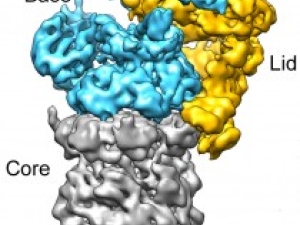
Berkeley researchers have provided the most detailed look ever at the “regulatory particle” used by the proteasome - one of the most critical protein machines in living cells - to identify and degrade proteins marked for destruction.