Agriculture Accelerated Human Genome Evolution To Capture Energy From Starchy Foods
UC Berkeley study finds rapid increase over last 12,000 years in genes for enzymes that digest starch.
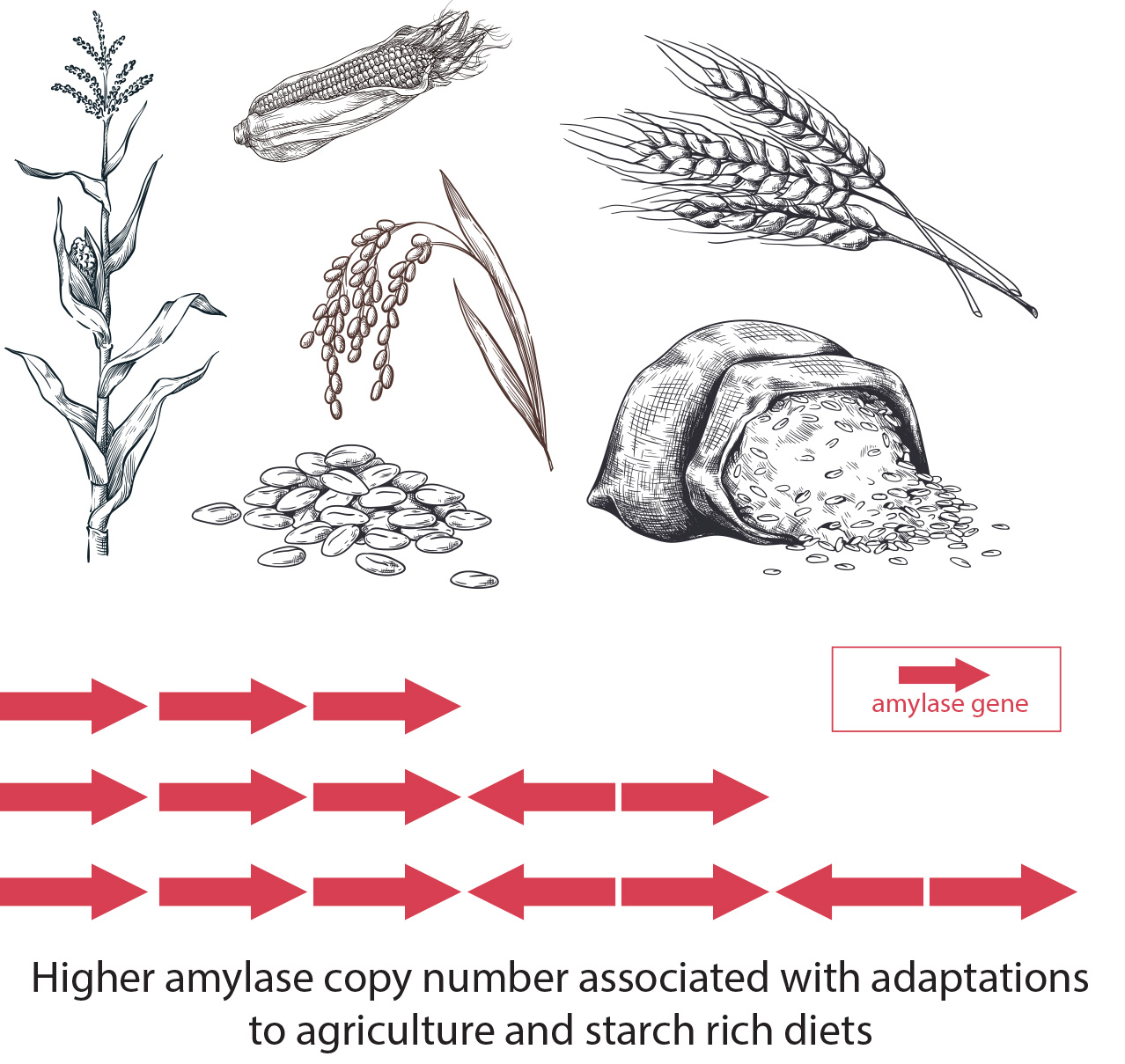
Over the past 12,000 years, humans in Europe have dramatically increased their ability to digest carbohydrates, expanding the number of genes they have for enzymes that break down starch from an average of eight to more than 11, according to a new study by researchers from the U.S., Italy and United Kingdom.
The rise in the number of genes that code for these enzymes tracks the spread of agriculture across Europe from the Middle East, and with it, an increasingly starchy human diet rich in high-carbohydrate staples such as wheat and other grains. Having more copies of a gene usually translates to higher levels of the protein the genes code for — in this case, the enzyme amylase, which is produced in saliva and the pancreas to break down starch into sugar to fuel the body.
The study, published today (Sept. 4) in the journal Nature, also provides a new method for identifying the causes of diseases that involve genes with multiple copies in the human genome, such as the genes for amylase.
The research was led by Peter Sudmant, assistant professor of integrative biology at the University of California, Berkeley, and Erik Garrison of the University of Tennessee Health Science Center in Memphis.
“If you take a piece of dry pasta and put it in your mouth, eventually it’ll get a little bit sweet,” Sudmant said. “That’s your salivary amylase enzyme breaking the starches down into sugars. That happens in all humans, as well as in other primates.”
Chimpanzee, bonobo and Neanderthal genomes all have a single copy of the gene on chromosome 1 that codes for the salivary amylase, referred to as AMY1. The same is true for the two pancreatic amylase genes, AMY2A and AMY2B. These three genes are located close to one another in a region of the primate genome known as the amylase locus.
Human genomes, however, harbor vastly different numbers of each amylase gene.
“Our study found that each copy of the human genome harbors one to 11 copies of AMY1, zero to three copies of AMY2A, and one to four copies of AMY2B,” said UC Berkeley postdoctoral fellow Runyang Nicolas Lou, one of five first authors of the paper. “Copy number is correlated with gene expression and protein level and thus the ability to digest starch.”
The researchers discovered that, while around 12,000 years ago humans across Europe had an average of about four copies of the salivary amylase gene, that number has increased to about seven. The combined number of copies of the two pancreatic amylase genes also increased by half a gene (0.5) on average over this time in Europe.
Survival advantage of multiple amylase genes
Overall, the incidence of chromosomes with multiple copies of amylase genes (that is, more total copies than chimpanzees and Neanderthals) increased sevenfold over the last 12,000 years, suggesting that this provided a survival advantage for our ancestors.
The researchers also found evidence for an increase in amylase genes in other agricultural populations around the world, and that the region of the chromosomes where these amylase genes are located looks similar in all these populations, no matter what specific starchy plant that culture domesticated. The findings demonstrate that as agriculture arose independently around the world, it seems to have rapidly altered the human genome in nearly identical ways in different populations to deal with increased carbohydrates in the diet.
In fact, the researchers found that the rate of evolution leading to changes in amylase gene copy number was 10,000 times faster than that of single DNA base pair changes in the human genome.
“It has long been hypothesized that the copy number of amylase genes had increased in Europeans since the dawn of agriculture, but we had never been able to sequence this locus fully before. It is extremely repetitive and complex,” Sudmant said. “Now, we’re finally able to fully capture these structurally complex regions, and with that, investigate the history of selection of the region, the timing of evolution and the diversity across worldwide populations. Now, we can start thinking about associations with human disease.”
One suspected association is with tooth decay. Previous studies have suggested that having more copies of AMY1 is associated with more cavities, perhaps because the saliva does a better job of converting starch in chewed food into sugar, which feeds bacteria that eat away at teeth.
The research also provides a method for exploring other areas of the genome — those involving the immune system, skin pigmentation and the production of mucus, for example — that have undergone rapid gene duplication in recent human history, Garrison said.
“One of the exciting things we were able to do here is probe both modern and ancient genomes to dissect the history of structural evolution at this locus,” he said.
These methods can also be applied to other species. Previous studies have shown that animals that hang out around humans — dogs, pigs, rats and mice — have more copies of the amylase gene than their wilder relatives, apparently to take advantage of the food we throw away.
“This is really the frontier, in my opinion,” Garrison said. “We can, for the first time, look at all of these regions that we could never look at before, and not just in humans — other species, too. Human disease studies have really struggled in identifying associations at complex loci, like amylase. Because the mutation rate is so high, traditional association methods can fail. We’re really excited how far we can push our new methods to identify new genetic causes of disease.”
From hunter-gatherer to agrarian
Scientists have long suspected that humans’ ability to digest starch may have increased after our ancestors transitioned from a hunter-gatherer lifestyle to a settled, agricultural lifestyle. This shift was shown to be associated with more copies of the amylase genes in people from societies that domesticated plants.
But the area of the human genome where these copies reside has been difficult to study because traditional sequencing — so-called short-read sequencing techniques that cut the genome into chunks of about 100 base pairs, sequence the millions of pieces and then reassemble them into a genome — was unable to distinguish gene copies from one another. Complicating matters, some copies are inverted, that is, they are flipped and read from the opposite strand of DNA.
Long-read sequencing allows scientists to resolve this region, reading DNA sequences thousands of base pairs long to accurately capture repetitive stretches. At the time of the study, the Human Pangenome Reference Consortium (HPRC) had collected long-read sequences of 94 human haploid genomes, which Sudmant and colleagues used to assess the variety of contemporary amylase regions, called haplotypes. The team then assessed the same region in 519 ancient European genomes. The HPRC data helped avoid a common bias in comparative genomic studies, which have used a single, averaged human genome as a reference. The genomes from the HPRC, referred to as a pangenome, provide a more inclusive reference that more accurately captures human diversity.
Joana Rocha, a UC Berkeley postdoctoral fellow and co-first author of the paper, compared the region where amylase genes cluster to what she called “sculptures made of different Lego bricks. Those are the haplotype structures. Previous work had to take down the sculpture first and infer from a pile of bricks what the sculpture may have looked like. Long-read sequencing and pangenomic methods now allow us to directly examine the sculpture and thus offer us unprecedented power to study the evolutionary history and selective impact of different haplotype structures.”
Using specially developed mathematical modeling, the researchers identified 28 different haplotype structures among the 94 long-read genomes and thousands of realigned short-read human genomes, all of which cluster into 11 groups, each with a unique combination of AMY1, AMY2A and AMY2B copy numbers.
“These remarkably complex, crazy structures — regions of gene duplication, inversion and deletion in the human genome — have evolved independently in different human populations over and over again, even before the rise of agriculture,” Sudmant said.
Analysis of the many contemporary human genomes also pointed to an origin 280,000 years ago of an initial duplication event that added two copies of AMY1 to the human genome.
“That particular structure, which is predisposed to high mutation rates, emerged 280,000 years ago, setting the stage for later on, when we developed agriculture, for people who had more copies to have increased fitness, and then for these copy numbers to be selected for,” Sudmant said. “Using our methods, for the first time we could really date the initial duplication event.”
Alma Halgren, a UC Berkeley graduate student in bioengineering, and Davide Bolognini and Alessandro Raveane of Human Technopole in Milan, Italy, are also first authors of the paper. Other co-authors are Andrea Guarracino of UTHSC, Nicole Soranzo of Human Technopole and the University of Cambridge in the United Kingdom, and Jason Chin of the Foundation for Biological Data Science in Belmont, California. Sudmant’s research is funded by the Institute of General Medical Sciences of the U.S. National Institutes of Health (R35GM142916).
RELATED INFORMATION
