Research Bio
Robert Tjian is Professor of Biochemistry, Biophysics and Structural Biology, the Li Ka Shing Chancellor's Chair in Biology, and a HHMI Investigator.
Biochemistry of Transcription and Chromatin Transactions: Over the past 30 years, the Tjian + Darzacq Group lab has identified, isolated and characterized a large number (~100) of essential drosophila and human transcription factors. These regulatory proteins include enhancer/promoter recognition factors, core RNA pol II initiation factors and co-activators that form large multi-subunit complexes at promoter DNA to mediate transcription initiation and decode the genome. Their studies indicate that large co-activator complexes play a critical role in mediating both universal as well as cell type specific networks of gene transcription and can serve as the interface between transcription and chromatin regulation. Although their earlier studies focused on in vitro biochemistry and conventional molecular genetics approaches, their recent research is directed at the more challenging task of integrating specific molecular transactions with in vivo mechanisms in cells and organisms under physiological conditions.
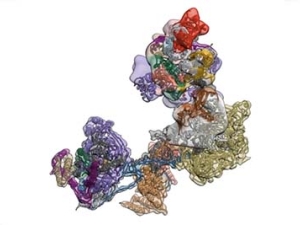
Single Cell Biochemistry, Structural Biology and Single Molecule Measurements: In order to address mechanisms of transcriptional control they use a combination of structural biological methods such as cryo EM/3D particle reconstruction, X-ray crystallography and single molecule measurements. Another effort is to develop strategies for measuring the assembly and movement of transcription complexes at the single cell level. These studies aim to track the movement and function of target transcription factors in live cells by various optical methods. In addition, they continue to utilize tools such as siRNA mediated knockdown of transcription factors as well as in vitro reconstitution of regulated transcription to link specific molecular mechanisms of control with in vivo physiology.
Transcription Mechanisms in Stem Cell Pluripotency and Differentiation: Deciphering the molecular basis of pluripotency is fundamental to our understanding of development and embryonic stem (ES) cell function. Using techniques such as ChIP-sequence, ATAC and Micro-C analysis, systematic RNAi mutagenesis, transgenic animals or cells, and in vitro transcription assays, they have developed complimentary experimental strategies to dissect in vivo mechanisms of transcriptional regulation and tease out how these various protein/DNA and protein/protein transactions influence gene expression. They recently found that single particle tracking reveals many aspects of biomolecular transactions in vivo such as their highly dynamic nature (residents times of only a few sec), anisotropic diffusion behavior and formation of local, highly transient protein “Hubs” via multivalent protein:protein interactions. These and related findings using “Fast SPT” are forcing us and the field to rethink many traditional and accepted models for how various processes and reactions such as transcription control actually take place in living cells. In addition, the lab is engaged in using these super-resolution imaging and genomics platforms to dissect disease mechanisms including cancer, viral infection, developmental disorders and metabolic imbalance. The lab is also continuing its efforts to develop new imaging tools, both hardware and software to more efficiently collect and process the vast amounts of data being collected using super-fast video.
Research Expertise and Interest
eukaryotic molecular biology; biochemistry, cellular differentiation, chromatin function, RNA synthesis, single cell imaging, single molecule imaging, disease mechanisms and translational drug therapy strategies including targeting, transcription factor activities with small molecule Glues via induced proximity and LockTac modalities
In the News
New Fellows of the American Association for the Advancement of Science
Freeze-frame microscopy captures molecule’s ‘lock-and-load’ on DNA
Scientists decipher opening dance steps of DNA and its partner
Working at temperatures near absolute zero, UC Berkeley experts in electron microscopy have learned in detail how proteins orchestrate the first key steps in gene activation – opening up the double-stranded DNA.