Thanks to rapid, 3D imaging, anyone can tour the fly brain
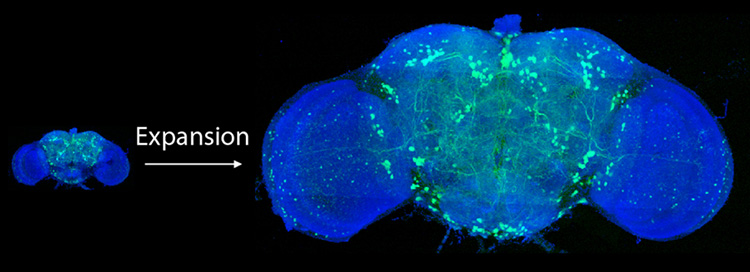
A new fly-through of the fly brain allows anyone to whizz past neurons and visit any of the 40 million synapses where neurons touch neuron. It’s a super-resolution view of the complex network connections in the insect’s brain that underlie behaviors ranging from feeding to mating.
What’s unprecedented, however, is that this 3D map over the whole fly brain, which shows details as small as 60 nanometers across, was captured in less than three days.
While the level of detail is not quite as good as that obtained with an electron microscope, efforts to fully map the neurons and synapses of the fly brain with EM have taken 10 years and the efforts of dozens of people. The new map was obtained a thousand times faster by combining two state-of-the-art techniques, expansion microscopy and lattice light-sheet microscopy.
A fine-scale map of the complete neural network of the brain – the human brain but also that of the mouse and fly – has been a dream of neuroscientists for decades. With it, they could trace the connections between neurons to understand how the brain makes decisions. And by counting synapses, neuroscientists could judge the strength of neural connections, such as those responsible for memory.
The new and faster whole-brain imaging technique will help scientists tease out the fly neural circuits that ultimately also underlie human brain functioning. And it works equally well for mapping the neural circuits in small chunks of the mouse brain, and potentially the human brain.
“You can spend years and years getting an EM image of one fly brain,” said Nobel laureate Eric Betzig, who invented the lattice light-sheet microscope while at the Janelia Research Campus of the Howard Hughes Medical Institute and is now a professor of molecular and cell biology and of physics at UC Berkeley. “I can see us getting to the point of imaging at least 10 fly brains per day.”
Such speed and resolution will let scientists ask new questions, he said, like how brains differ between males and females, or how brain circuits vary between flies of the same type.
“We’ve crossed a threshold in imaging performance,” said Edward Boyden of the Massachusetts Institute of Technology, who invented expansion microscopy five years ago. “That’s why we’re so excited. We’re not just scanning incrementally more brain tissue, we’re scanning entire brains.”
Betzig, Boyden and their teams will publish their findings this week in the journal Science.
Combining expansion and light-sheet microscopy
The new brain-scanning technique came about after Boyden asked Betzig’s help in combining expansion microscopy with Betzig’s latest high-speed imaging technique, lattice light-sheet microscopy. Expansion microscopy (ExM) involves fixing tissue and then expanding it like a balloon while keeping the relative positions of internal structures unchanged. It uses a polyacrylamide gel like that in diapers, which swells when moved from salty to pure water. Lattice light-sheet microscopy (LLSM) uses highly focused light beams to rapidly assemble a 3D image of a specimen one thin slice at a time.
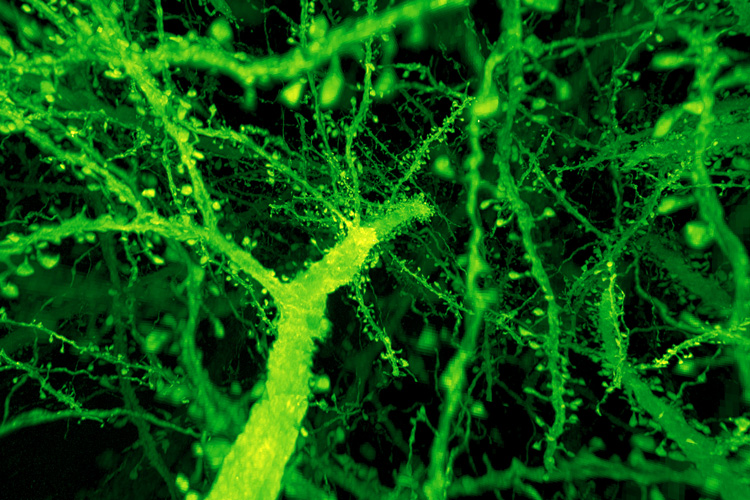
“When they first came to me in 2016, I was still a skeptic; I was worried, first, whether you could expand anything like that and not have it be warped like crazy,” Betzig said. “And then I was afraid that, while the samples are transparent, they would still warp the light like a bag of marbles.”
Working at the Janelia campus, the combined teams, led by postdocs Ruixuan Gao and Shoh Asano of Boyden’s MIT lab and Srigokul Upadhyayula of Harvard Medical School, found that after expanding the brain tissue by a factor of four, to a volume 64 times normal, it was nearly as clear as water and unwarped.
“I was shocked at how perfect was the clearing to make it incredibly optically uniform,” he said.
As a result, the lattice light-sheet microscope was able to produce a highly detailed and precise image of the brain at the level of single synapses: a resolution of about 60 nanometers, which is only one-tenth the resolution of EM. The multicolor imaging took just 62.5 hours.
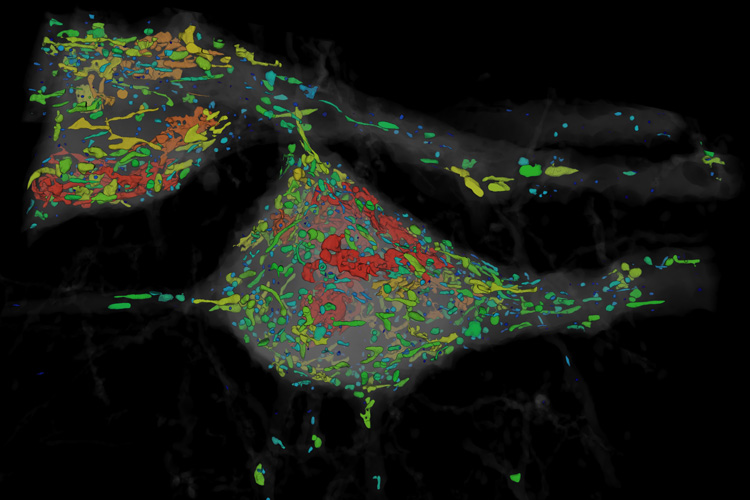
The teams tested the ExLLSM technique not only on the entire fruit fly brain, but also on a sliver of mouse brain spanning the millimeter-thick cortex, with similar results. They were able to count all the synapses in the fly brain, totaling some 40 million. The human brain, with 80 billion neurons and perhaps 7,000 synapses per neuron, would be much more challenging.
Betzig predicts, however, that with improvements in expansion microscopy — some scientists have stretched tissue 25 times in each direction — the combined techniques could achieve results nearly as good as EM in terms of mapping all the neural connections in the brain, a field known as connectomics.
“If you could get it to work at 10 times or maybe 15 times expansion, you could probably put a lot of the EM out of business,” he said. “It might be good enough to do the dense neural tracing EM can do but much much faster and cheaper. I think they need to watch their backs. It is not there yet, but in my opinion, the potential is there.”
Fluorescence microscopy
Both microscope techniques involve labeling proteins in the tissue with fluorescent markers. In expansion microscopy, the tissue is then infused with the gel and the markers cross-linked to the gel structure. Then all the protein is digested away, leaving what Betzig terms a “fluorescent phantom.” Changing the salt concentration of the media causes the gel to swell, dragging the markers with it. It becomes mostly water, which accounts for its clarity.
Boyden originally used confocal light microscopy to image the expanded tissue, but he hoped that LLSM would image the sample faster and with even better resolution, while also overcoming the complete loss of fluorescence signal that occurs when imaging deep within thick specimens by conventional means. LLSM scans a sheet of light as narrow as 400 nanometers, plane by plane through the sample, imaging the fluorescence from the markers in each illuminated plane. Each complete scan, amounting to nearly 10 terabytes of data, is then assembled by computer into a 3D image that can be navigated like a video game. The time-intensive assembly was overseen by Janelia computer scientists Stefan Saalfeld and Igor Pisarev, and then analyzed and visualized by Upadhyayula, who will soon open a state-of-the-art imaging lab at UC Berkeley.
By attaching fluorescent markers to any of the 10,000 proteins in the brain, it should be possible to map the outer membranes of neurons and other cells, the synapses where one neuron connects with another, the internal compartments of brain cells, and much more.
There are limitations, however. As with any kind of super-resolution fluorescence microscopy, Betzig said, it can be hard to decorate proteins with enough fluorescent bulbs to see them clearly at high resolution. And since expansion microscopy requires many processing steps, there’s still the potential for artifacts to be introduced. Because of this, he said, “we worked very hard to validate what we’ve done, and others would be well-advised to do the same.”
The work was funded primarily by HHMI and the National Institutes of Health.
RELATED INFORMATION
