So-Called Junk DNA Plays Critical Role in Mammalian Development

Nearly half of our DNA has been written off as junk, the discards of evolution: sidelined or broken genes, viruses that got stuck in our genome and were dismembered or silenced, none of it relevant to the human organism or human evolution.
But research over the last decade has shown that some of this genetic “dark matter” does have a function, primarily in regulating the expression of host genes — a mere 2% of our total genome — that code for proteins. Biologists continue to debate, however, whether these regulatory sequences of DNA play essential or detrimental roles in the body or are merely incidental, an accident that the organism can live without.
A new study led by researchers at University of California, Berkeley, and Washington University explored the function of one component of this junk DNA, transposons, which are selfish DNA sequences able to invade their host genome. The study shows that at least one family of transposons — ancient viruses that have invaded our genome by the millions — plays a critical role in viability in the mouse, and perhaps in all mammals. When the researchers knocked out a specific transposon in mice, half their mouse pups died before birth.
This is the first example of a piece of “junk DNA” being critical to survival in mammals.
In mice, this transposon regulates the proliferation of cells in the early fertilized embryo and the timing of implantation in the mother’s uterus. The researchers looked in seven other mammalian species, including humans, and also found virus-derived regulatory elements linked to cell proliferation and timing of embryo implantation, suggesting that ancient viral DNA has been domesticated independently to play a crucial role in early embryonic development in all mammals.
According to senior author Lin He, UC Berkeley professor of molecular and cell biology, the findings highlight an oft-ignored driver of evolution: viruses that integrate into our genome and get repurposed as regulators of host genes, opening up evolutionary options not available before.
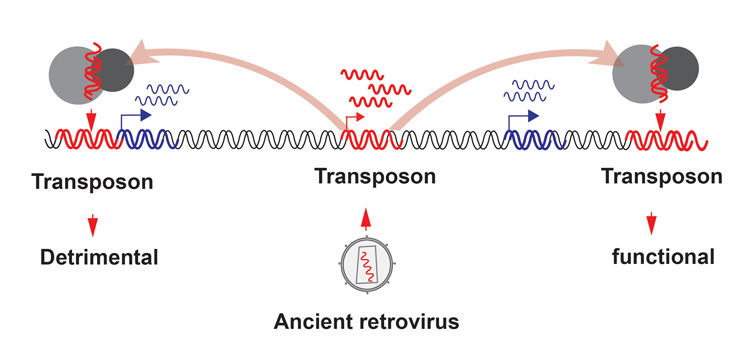
“The mouse and humans share 99% of their protein coding genes in their genomes — we are very similar with each other,” He said. “So, what constitutes the differences between mice and humans? One of the major differences is gene regulation — mice and humans have the same genes, but they can be regulated differently. Transposons have the capacity to generate a lot of gene regulatory diversity and could help us to understand species-specific differences in the world.”
Colleague and co-senior author Ting Wang, the Sanford and Karen Loewentheil Distinguished Professor of Medicine in the Department of Genetics at the Washington University School of Medicine in St. Louis, Missouri, agrees.
“The real significance of this story is it tells us how evolution works in the most unexpected manner possible,” Wang said. “Transposons were long considered useless genetic material, but they make up such a big portion of the mammalian genome. A lot of interesting studies illustrate that transposons are a driving force of human genome evolution. Yet, this is the first example that I know of where deletion of a piece of junk DNA leads to a lethal phenotype, demonstrating that the function of specific transposons can be essential.”
The finding could have implications for human infertility. According to first author Andrew Modzelewski, a UC Berkeley postdoctoral fellow, nearly half of all miscarriages in humans are undiagnosed or don’t have a clear genetic component. Could transposons like this be involved?
“If 50% of our genome is non-coding or repetitive — this dark matter — it is very tempting to ask the question whether or not human reproduction and the causes of human infertility can be explained by junk DNA sequences,” he said.
Embryo implantation
He, the Thomas and Stacey Siebel Distinguished Chair Professor at UC Berkeley, studies the 98% or more of our genome that does not code for proteins. For most of He’s career, she has focused on microRNAs and longer pieces of non-coding RNAs, both of which are potent gene regulators. Five years ago, however, her team accidentally discovered a microRNA regulator for a transposon family called MERVL (mouse endogenous retroviral elements) that was involved in cell fate determination of early mouse embryos. The unexpected abundance of transposon transcription in mouse embryos led He’s team to investigate the developmental functions of transposons, which have taken up residence in the genomes of nearly every organism on Earth.

In a paper appearing last week in the journal Cell, He and her team identify the key regulatory DNA involved: a piece of a transposon — a viral promoter — that has been repurposed as a promoter for a mouse gene that produces a protein involved in cell proliferation in the developing embryo and in the timing of implantation of the embryo. A promoter is a short DNA sequence that is needed upstream of a gene in order for the gene to be transcribed and expressed.
Wild mice use this transposon promoter, called MT2B2, to initiate transcription of the gene Cdk2ap1 specifically in early embryos to produce a short protein “isoform” that increases cell proliferation in the fertilized embryo and speeds its implantation in the uterus. Using CRISPR-EZ, a simple and inexpensive technique that Modzelewski and He developed several years ago, they disabled the MT2B2 promoter and found that mice instead expressed the Cdk2ap1 gene from its default promoter as a longer form of the protein, a long isoform, that had the opposite effect: decreased cell proliferation and delayed implantation.
The result of this knockout was the death at birth of about half the pups.
Modzelewski said that the short form of the protein appears to make the many embryos of the mouse implant with a regular spacing within the uterus, preventing crowding. When the promoter is knocked out so that the long form is present only, the embryos implant seemingly randomly, some of them over the cervix, which blocks exit of the fully developed fetus and sometimes kills the mother during the birthing process.
They found that within a 24-hour period prior to embryo implantation, the MT2B2 promoter ramps up expression of the Cdk2ap1 gene so much that the short form of the protein makes up 95% of the two isoforms present in embryos. The long isoform is normally produced later in gestation when the default promoter upstream of the Cdk2ap1 gene becomes active.
Working with Wanqing Shao, co-first author of the study and a postdoctoral fellow in Wang’s group at Washington University, the team searched through published data on preimplantation embryos for eight mammalian species — human, rhesus monkey, marmoset, mouse, goat, cow, pig and opossum — to see whether transposons are turned on briefly before implantation in other species. These online data came from a technique called single cell RNA sequencing, or scRNA-seq, which records the levels of messenger RNA in single cells, an indication of which genes are turned on and transcribed. In all cases, they had to retrieve the data on non-coding DNA because it is typically removed before analysis, with the presumption that it’s unimportant.
While transposons are generally specific to individual species — humans and mice, for example, have largely different sets — the researchers found that different species-specific transposon families were turned on briefly before implantation in all eight mammals, including the opossum, the only mammal in the group that does not employ a placenta to implant embryos in the uterus.
“What’s amazing is that different species have largely different transposons that are expressed in preimplantation embryos, but the global expression profiles of these transposons are nearly identical among all the mammalian species,” He said.
Colleague and co-senior author Davide Risso, a former UC Berkeley postdoctoral fellow and now associate professor of statistics at the University of Padua in Italy, developed a method for linking specific transposons to preimplantation genes so as to weed out the thousands of copies of related transposons that exist in the genome. This method is crucial to identifying individual transposon elements with important gene regulatory activity.
“It’s interesting to note that the data that we used were mostly based on the previous sequencing technology, called SMART-seq, which covers the full sequence of the RNA molecules. The current popular technique, 10x genomics technology, would not have shown us the different levels of protein isoforms. They’re blind to them,” Risso said.
Viruses are evolutionary reservoir
The researchers found that in nearly all of the eight mammalian species, both short and long Cdk2ap1 isoforms occur, but are switched on at different times and in different proportions that correlate with whether embryos implant early, as in mice, or late, as in cows and pigs. Thus, at the protein level, both the short and long isoforms appear conserved, but their expression patterns are species-specific.
“If you have a lot of the short Cdk2ap1 isoform, like mice, you implant very early, while in species like the cow and pig, which have none to very little of the short isoform, it’s up to two weeks or longer for implantation,” Modzelewski said.
Wang suspects that the promoter that generates the long form of the protein could be the mouse’s original promoter, but that a virus that integrated into the genome long ago was later adapted as a regulatory element to produce the shorter form and the opposite effect.
“So, what happened here is a rodent-specific virus came in, and then somehow the host decided, ‘OK, I’m going to use you as my promoter to express this shorter Cdk2ap1 isoform.’ We see the redundancy that’s built into the system, where we can take advantage of whatever nature throws at us and make it useful,” he said. “And then, this new promoter happened to be stronger than the old promoter. I think this fundamentally changed the phenotype of rodents; maybe that’s what makes them grow faster — a gift of having a shorter pre-implantation time. So, they probably gained some fitness benefit from this virus.”
“Whatever you look at in biology, you’re going to see transposons being used, simply because there are just so many sequences,” Wang added. “They essentially provide an evolutionary reservoir for selection to act upon.”
Other co-authors of the study are Jingqi Chen, Angus Lee, Xin Qi, Mackenzie Noon, Kristy Tjokro and Anne Biton of UC Berkeley; Terry Speed of the Walter and Eliza Hall Institute of Medical Research in Melbourne, Australia; Aparna Anand of Washington University and Gabriele Sales of the University of Padua. The work was supported primarily by the Howard Hughes Medical Institute faculty scholar award and the National Institutes of Health.
RELATED INFORMATION
