Scientists reprogram embryonic stem cells to expand their potential
Researchers from UC Berkeley have found a way to reprogram mouse embryonic stem cells so that they exhibit developmental characteristics resembling those of fertilized eggs, or zygotes.
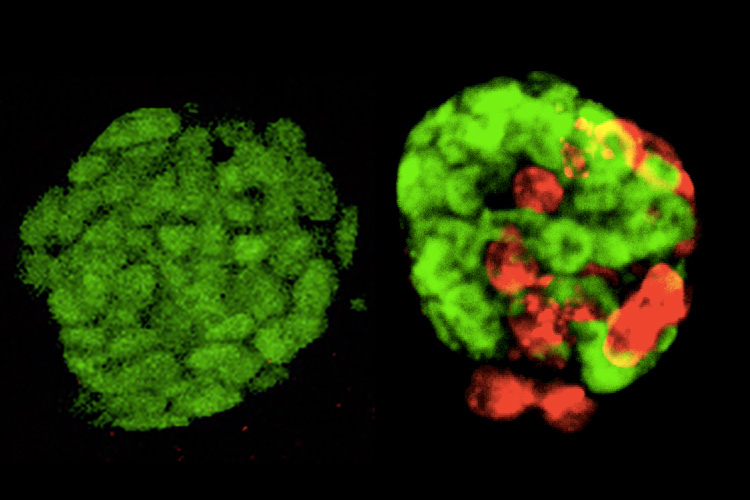
These “totipotent-like” stem cells are able to generate not only all cell types within a developing embryo, but also cell types that facilitate nutrient exchange between the embryo and the mother.
For now, the new stem cell lines UC Berkeley researchers have created will help scientists understand the first molecular decisions made in the early embryo. Ultimately, however, these insights could broaden the repertoire of tissues that can be generated from stem cells, with significant implications for regenerative medicine and stem cell-based therapy.
A fertilized egg is thought to possess full developmental potential, able to generate all cell types required for embryo gestation, including the developing embryo and its extra-embryonic tissues. A unique feature of placental mammals, extra-embryonic tissues such as the placenta and yolk sac are vital for nutrient and waste exchange between the fetus and mother.
By contrast, most embryonic and induced pluripotent stem cells are more restricted in their developmental potential, able to form embryonic cell types, but not extra-embryonic tissues. The ability of a fertilized egg to generate both embryonic and extra-embryonic tissues is referred to as “totipotency,” an ultimate stem cell state seen only during the earliest stages of embryonic development.
“Studies on embryonic development greatly benefit from the culture system of embryonic stem cells and, more recently, induced pluripotent stem cells. These experimental systems allow scientists to dissect key molecular pathways that specify cell fate decisions in embryonic development,” said team leader Lin He, a UC Berkeley associate professor of molecular and cell biology. “But the unique developmental potential of a zygote, formed right after the sperm and egg meet, is very, very difficult to study, due to limited materials and the lack of a cell-culture experimental system.”
He’s new study not only reveals a novel mechanism regulating the “totipotent-like” stem cell state, but also provides a powerful cell-culture system to further study totipotency.
She and her colleagues reported their research online Jan. 12 in advance of print publication in the journal Science.
MicroRNAs and stem cells
Embryonic stem (ES) cells, harvested from three-and-a-half-day-old mouse embryos or five-and-a-half-day-old human embryos, are referred to as pluripotent because they can become any of the thousands of cell types in the body. They have generated excitement over the past few decades because scientists can study them in the laboratory to discover the genetic switches that control the development of specialized tissues in the embryo and fetus, and also because of their potential to replace body tissues that have broken down, such as pancreatic cells in those with diabetes or heart muscle cells in those with congestive heart failure. These stem cells can also let researchers study the early stages of genetic disease.
As an alternative to harvesting them from embryos, scientists can also obtain pluripotent stem cells by treating mature somatic cells with a cocktail of transcription factors to regress them so that they are nearly as flexible as embryonic stem cells. These artificially derived stem cells are called induced pluripotent stem (iPS) cells.
Neither ES nor iPS cells, however, are as flexible as the original fertilized egg, which can form extra-embryonic as well as embryonic tissues. By the time embryonic stem cells are harvested from a mouse or human embryo, the cells have already committed to either an embryonic or an extra-embryonic lineage.
MicroRNAs are small, non-coding RNAs that do not translate into proteins, yet have a profound impact on gene expression regulation. He and her colleagues found that a microRNA called miR-34a appears to be a brake preventing both ES and iPS cells from producing extra-embryonic tissues. When this microRNA was genetically removed, both ES and iPS cells were able to expand their developmental decisions to generate embryo cell types as well as placenta and yolk sac linages. In their experiments, about 20 percent of embryonic stem cells lacking the microRNA exhibited expanded fate potential. Furthermore, this effect could be maintained for up to a month in cell culture.
“What is quite amazing is that manipulating just a single microRNA was able to greatly expand cell fate decisions of embryonic stem cells,” He said. “This finding not only identifies a new mechanism that regulates totipotent stem cells, but also reveals the importance of non-coding RNAs in stem cell fate.”
Additionally, in this study, He’s group discovered an unexpected link between miR-34a and a specific class of mouse retrotransposons. Long regarded as “junk DNA,” retrotransposons are pieces of ancient foreign DNA that make up a large fraction of the mammalian genome. For decades, biologists assumed that these retrotransposons serve no purpose during normal development, but He’s findings suggest they may be closely tied to the decision-making of early embryos.
“An important open question is whether these retrotransposons are real drivers of developmental decision making,” said Todd MacFanlan, a co-author of the current study and a researcher at the Eunice Kennedy Shriver National Institute of Child Health and Human Development in Bethesda, Maryland.
Co-authors with He are graduate student Yong Jin Choi, postdoctoral fellows Chao-Po Lin and Davide Risso, graduate student Sean Chen and undergraduate Thomas Aquinas Kim, along with statistics professor Terence Speed of UC Berkeley. Meng How Tan and Jin Li of Stanford University, Yalei Wu of Thermo Fisher Scientific in South San Francisco, Caifu Chen of Integrated DNA Technologies in Redwood City, Zhenyu Xuan of the University of Texas at Dallas, Weiqun Peng of George Washington University in Washington, D.C., Kent Lloyd of UC Davis and Sang Yong Kim of the New York University School of Medicine, all contributed to this work.
He’s work was funded by the California Institute for Regenerative Medicine (RN2-00923-1), National Cancer Institute (R01 CA139067), National Institute of General Medical Sciences (R01GM114414) and a faculty scholar award from the Howard Hughes Medical Institute.
RELATED INFORMATION
