Reconstructing the Chromosomes of the Earliest Animals on Earth

Many of today’s marine invertebrates, including sponges and jellyfish, have chromosomes with the same ancient structure they inherited from their primitive ancestors more than 600 million years ago, according to a new study.
The surprise finding is a reminder that evolution is conservative — it keeps things that work well, like the organization of genes on a chromosome — and provides a key link between creatures alive today, including humans, and our very distant ancestors.
“It emphasizes that even in something as fundamental as their chromosomes, diverse animals resemble each other,” said the study’s senior author, Daniel Rokhsar, the Marthella Foskett Brown Chair in the Department of Molecular and Cell Biology at the University of California, Berkeley. “That’s one of the reasons why we can learn so much about human biology from studying fruit flies, nematode worms, jellyfish and other ‘simple’ model systems — it’s because of the underlying unity of all animals. What we learn about animal diversity affects how we think about ourselves.”
The findings were published Feb. 2 in the journal Science Advances.

The new analysis predicts that the first multicellular animals carried their genes in 29 pairs of ancient chromosomal units. As the first animals arose in the oceans and evolved into diverse invertebrates, from sponges to worms to humans, many of these chromosomes have remained intact for half a billion years.
For comparison, humans now have 23 pairs of chromosomes, for a total of 46, the result of two duplications and multiple mergers and chromosomal rearrangements since the earliest animals.
The study, led by Rokhsar and Oleg Simakov of the University of Vienna in Austria, is the first to compare the chromosomal position of genes from diverse animals, such as sponges, jellyfish, sea scallops and other aquatic invertebrates, allowing the ancestral organization to be inferred and rare changes in chromosome organization to be studied. Though this kind of analysis has been done for fruit flies and many vertebrates, including humans, it is only recently that the chromosome-scale genomes of diverse invertebrates have been determined.
Evolution is conservative
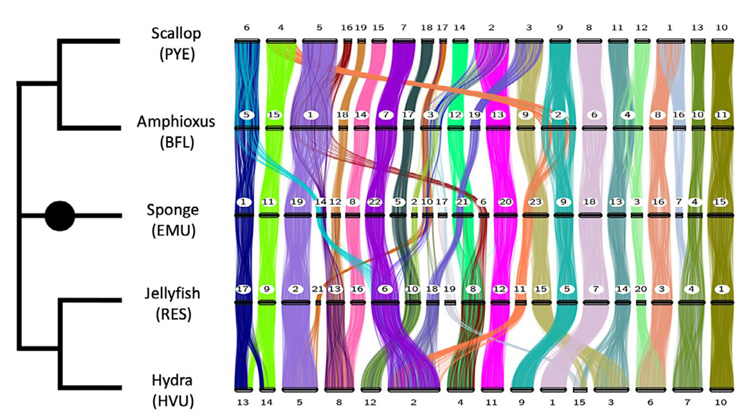
Because of increasingly advanced techniques for identifying which genes are close to one another when the chromosome is curled up inside the nucleus, scientists over the past few years have begun assigning genes to chromosomes in several invertebrates: the Florida lancelet, Branchiostoma floridae, a dainty, quill-like sea creature also known as amphioxus; a scallop, Patinopecten yessoensis; a fresh water sponge, Ephydatia muelleri; and the fire jellyfish, Rhopilema esculentum, a cnidarian. Rokhsar, Simakov and their team extended this set by determining the chromosomal sequences of a fifth animal, a hydra, Hydra vulgaris, another type of cnidarian.

“What we find is remarkable: If you compare those five species with each other, you find that there’s extensive conservation; in many cases, whole chromosomes or big pieces of chromosomes have stayed together. A whole chromosome in a sponge might correspond to a chromosome in a jellyfish,” he said. “They’re not organized in exactly the same way — the genes are in a different order in the various species — but over these long-time scales, a chromosome behaves like a bag of genes that has maintained its integrity since the beginning of animal life in the pre-Cambrian era.”
Once they discovered, in their sample of invertebrates, that genes tended to remain together on the same chromosome — something referred to as synteny, from the Greek for “on the same thread” — they predicted that the same would be true of other invertebrates, including sea urchins and various kinds of worms and mollusks. When they looked at the chromosomes of these organisms, they found similar conservation of DNA across chromosomes. All seemed to harken back to the same 29 chromosomal pairs that were present in the early animal ancestors.
What does this mean for humans and other vertebrates?

“If you compare amphioxus to scallops and then representatives of a lot of different vertebrates — different kinds of fish, like lampreys, chickens, and so forth — you can see that there are 18 different groups of genes that seem to always stick together,” said Rokhsar, who is also a Chan Zuckerberg Biohub investigator and a member of the Joint Genome Institute at the Lawrence Berkeley National Laboratory. “They always travel together on the same piece of DNA, and so the simplest interpretation is that there were 18 ancestral chromosomes in the proto-vertebrate ancestor.”
Rokhsar and his team have long suspected that chromosomes were more preserved than people thought. Over the past 20 years, he and his group have sequenced and analyzed the genomes of diverse animals, including a sea squirt, a placozoan, a species of lancelet and a different species each of sponge, choanoflagellate, sea anemone, octopus, acorn worm, leech, limpet and polychaete worm. While the early “draft” genomes were often fragmented, they nevertheless showed signs that there were anciently conserved groups of genes linked together across diverse animals. Newer technologies that allow whole chromosomes to be determined have confirmed those early hypotheses.

The fact that the genes of diverse invertebrates group together so faithfully, despite hundreds of millions of years of independent evolution, could indicate that for genes to jump around among chromosomes is a lot harder than scientists presumed from their studies of vertebrates, where genes have rearranged more frequently, likely because of genetic drift.
“Animals like amphioxus live in huge populations where the rare mutants with rearranged chromosomes are at a disadvantage and typically die out, whereas, in small, subdivided populations, which is more typical of mammals, rearrangements are more likely to survive and spread. That’s one hypothesis,” said Rokhsar.
Vertebrates mixed it up
Alternatively, there may be some unknown reason why sets of genes have to remain together. One famous example is the Hox genes, which determine which end of the animal embryo forms the head and which the tail, and all gradations in between. These genes are all clustered together on one chromosome in most invertebrates, and this clustering is important for their deployment during development. The functional clustering of these genes may be an exception, however, and there’s no evidence yet that the clusters found in the recent study are functionally related, Rokhsar said.
The simple conservation of chromosomes stops with invertebrates, because early in vertebrate evolution, the entire genome was duplicated twice in the lineage leading to jawed vertebrates, a group that includes mammals, birds, reptiles, amphibians and most fish. During the course of these large-scale duplications, a series of chromosomal reorganizations forged the genomes of the earliest jawed vertebrates, which eventually gave rise to humans. By tracking groups of genes as they moved from one chromosome to another as the earliest vertebrates evolved, however, Rokhsar and collaborators were able to leap over the vertebrate-invertebrate divide and connect the earliest animal chromosomes with those of contemporary vertebrates.
“One of the cool things is that once we infer these ancient proto-chromosomes and organize them on the tree of life, then we can make predictions. If you go and sequence some other genomes, we predict that you will inevitably find that these genes are mixed together on the same chromosome,” he said. “Unlike physics or chemistry, you don’t usually get to make such predictions in biology. But now we know something, in a sense, about almost all animal genomes from this comparison.”
The work was supported by the National Institutes of Health (RO1 HD080708), the Chan Zuckerberg Biohub, and the Molecular Genetics Unit of the Okinawa Institute of Science and Technology Graduate University (OIST) in Japan, where Rokhsar has a joint appointment as a visiting professor.
Other co-authors of the paper are Jessen Bredeson, Kodiak Berkoff and Therese Mitros of UC Berkeley; Ferdinand Marletaz of OIST and University College in the U.K.; Darrin Schultz of UC Santa Cruz and the Monterey Bay Aquarium Research Institute; Brendan O’Connell and Richard Green of UC Santa Cruz; the late Paul Dear of Mote Research Ltd. in the U.K.; Daniel Martinez of Pomona College; Robert Steele of UC Irvine; and Charles David of the Ludwig Maximilian University of Munich in Germany.
