Microbes in the preemie gut
Scientists have for the first time sequenced and reconstructed the genomes of most of the microbes in the gut of a premature newborn and documented how the microbe populations changed over time.
Further studies involving more infants could eventually help researchers understand the causes of various intestinal problems that afflict preemies, in particular the sometimes fatal necrotizing enterocolitis, according to researchers at the University of California, Berkeley, the University of Pittsburgh School of Medicine and Stanford University. One unresolved question is whether these illnesses are caused by pathogenic strains of bacteria or just an imbalance in the microbe populations in the gut.
The study was posted online Dec. 29 in advance of print publication in the journal Proceedings of the National Academy of Sciences.
While this is not the first time that microbes in the human intestinal tract have been sequenced as a community, this is the first comprehensive look at a time series documenting colonization of the gut of a premature newborn, and one of few completely assembled community genomic datasets, said Jill Banfield, a UC Berkeley professor of earth and planetary science and of environmental science, policy and management.
“Sequencing of microbial communities has become exceedingly common, but many researchers work with essentially unassembled data and often analyze very short contiguous DNA sequences – genome fragments,” she said. “We actually go in and work out where the assemblies failed and fix them – what’s called curating the data – so we can build very complete genomes for most of the microbes.”
From acid mines to the preemie gut
Pediatric surgeon Michael J. Morowitz, until recently at The University of Chicago Medical Center but now with Children’s Hospital of Pittsburgh of the University of Pittsburgh Medical Center and an assistant professor of surgery at the University of Pittsburgh School of Medicine, first approached Banfield because of her pioneering work over the past decade sequencing microbial communities in extreme environments, such as the acid drainage from underground mines. He suggested that she tackle a unique human environment, the newborn intestinal tract. Unlike the adult gut, which may contain a couple of thousand microbial species, the newborn’s intestinal tract may be colonized by only a handful, making it feasible to sequence the entire community.
His interest stemmed from work with premature infants, most of whom spend anywhere from two weeks to six months in the intensive care unit before they’re deemed healthy enough to go home. Between 5 and 10 percent of these preemies develop symptoms of necrotizing enterocolitis (NEC), which requires rounds of antibiotics to halt, and perhaps a third of these babies eventually require surgery to remove parts of their intestines that have died.
“The actual impact of necrotizing enterocolitis in the ICU is even larger, because feeding routines and other care are conducted around a fear of NEC developing,” Morowitz said.
Previous studies, however, have produced conflicting results about NEC’s cause. Some have found pathogenic bacteria associated with NEC, while others have found no difference between the bacteria in babies with and without NEC. Banfield, Morowitz and their collaborators suspect that these results reflect the fact that researchers have looked broadly at species or families of bacteria in the gut, rather than at variants or strains. Although coexisting strains may have genes that are 99 percent identical, their genomes could be sufficiently distinct to make one bad and the other good.
“We already know that just a few genes can make one strain a pathogen and one beneficial or commensal,” meaning that the microbes live amicably with their host, Banfield said. “We expect that a lot of the issues with the colonization process in the gut that leads to disease may be tracked to subtle differences in strains,” she said. “So one question on the table is, ‘Are these very closely related strains physiologically distinct, and in what ways’?”
The only way to get at these differences, she said, is to sequence the entire genomes of the intestinal microbiota – not merely DNA fragments or short DNA tags, which can be used to identify the genus or even species of a microbe, but not the specific strain.
Colonizing the gut
“Although a primary target of our research is NEC, it’s become very apparent that there are some fundamental unanswered questions just about the colonization process under normal circumstances,” Morowitz added. “It’s really important to get a handle on what the normal process is first, and then, eventually, we can look closely at babies with NEC and see if they deviate from what appears to be the normal colonization process.”
Other human diseases, including asthma, diabetes and obesity, have been linked to problems with microbial colonization of the gut, and several papers have reported symptomatic improvement after “transplanting” fecal material from healthy individuals to patients with a range of intestinal disorders.
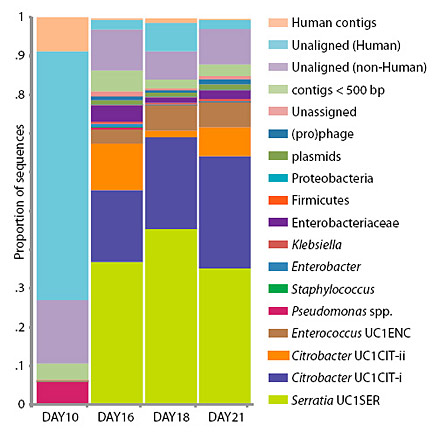
Banfield, Morowitz and their colleagues followed a single premature infant that had been delivered by cesarean and identified three distinct communities of intestinal microbes present at different times during the first month of the infant’s life. The microbe populations in these communities seemed to change after alterations in medication and feeding, Morowitz said. Although it was presumably sterile at birth, the infant’s gut was quickly colonized by a set of known intestinal microbes – bacteria and Archaea, primarily, but also viruses, bacterial viruses (phage) and the naked lengths of DNA called plasmids. When the baby went off antibiotics and switched from breast feeding to intravenous feeding, the microbe populations completely changed, with minor microbial members suddenly dominating and dominant members declining.
Vincent Denef, a post-doctoral researcher in Banfield’s lab who contributed to the study, noted that such real-time studies are powerful because “very rarely do we have the opportunity to observe the dynamics of a naturally occurring system, such as the infant GI tract, as it is transformed from sterile to functionally diverse.”
The populations again shifted when intravenous feeding was replaced by formula. Morowitz stopped collecting feces from dirty diapers after 21 days, and the infant was sent home healthy after 9 weeks in the ICU.
Though fecal samples were taken nearly every day, a complete genome analysis was performed only for samples collected on days 10, 16, 18 and 21. For the other days, the microbial community was estimated based on DNA tags (16S rRNA) that identify microbe families and species, but not specific strains.
What distinguishes bad from good microbes?
What surprised the researchers is that the microbial population was comprised of members of at least 20 groups, many of which include harmful as well as benign organisms. These included Staphylococcus, a frequent cause of hospital infections; Pseudomonas, “the cause of an enormous amount of morbidity in ICU patients, both children and adults,” Morowitz said; Serratia, a common cause of sepsis in general; and Citrobacter, which can cause meningitis in babies. Yet, the baby in this study appeared healthy throughout.
“The gut populations are highly dynamic, with large shifts through three stages over time, but we saw an overabundance of gram-negative organisms that we often associate with disease,” he said. “Particularly striking was the dominance of Pseudomonas for several days, though the infant was clinically stable.”
The seeming contradiction of a healthy infant with disease-causing bacteria in her gut could be explained if the strains in the infant’s gut were benign, or if the balance of other microbes prevented pathogenic microbes from causing problems.
Citrobacter, for example, is one type of bacteria that is reportedly associated with NEC: one study found Citrobacter in three of four infants with NEC, but in no control infants. Yet, in the current study, sequencing of the gut microbiome on days 16, 18 and 21 revealed the presence of two strains of Citrobacter, which fluctuated significantly in proportions on the three days. “Those big shifts could potentially have been very important for the medical state of that baby,” Banfield said. “Fortunately, the baby was fine.”
The researchers found that those two strains were 99 percent similar over areas of the genome that could be compared.
“Of particular interest were hot spots of rapid DNA evolution within and between genes. Those potentially could be very important and interesting,” she said. “Though the two Citrobacter genotypes are very, very similar over most of the genome, the results suggest that they could be functioning in different ways because their genomes are regulated differently.”
Banfield noted that the intestinal community of the infant no doubt would continue to shift repeatedly for a year or more after birth, as the child encounters new microbes – courtesy of family, friends and pets. These populations shift with the influx of new strains and species and potentially because the resident microbes themselves evolve by picking up new traits from the plasmids and phages living alongside them.
“This is an ecological study,” she emphasized. “One of the things we are trying to do is bring into the field of medicine a high resolution, ecological approach.”
Other coauthors of the study are Brian C. Thomas of UC Berkeley, Valeriy Poroykoa of the University of Chicago Pritzker School of Medicine, and David A. Relman and Elizabeth K. Costello of the Stanford University School of Medicine.
The work was funded by National Institute of Allergy and Infectious Diseases of the National Institutes of Health, the Department of Energy, the Surgical Infection Society and The March of Dimes.
For more information:
Link to PNAS paper
