Long-sought structure of telomerase paves way for drugs for aging, cancer
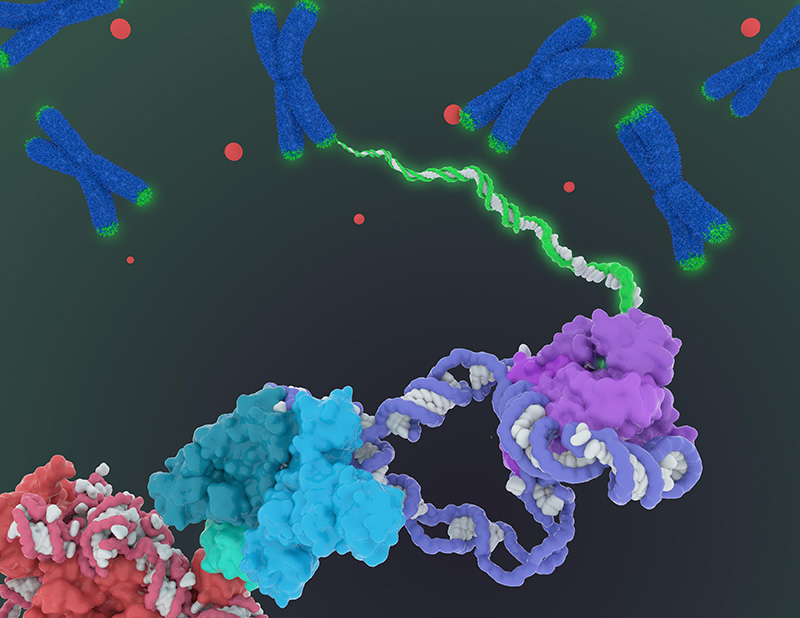
More than 30 years ago, when UC Berkeley researchers discovered telomerase — an enzyme that lengthens chromosome ends and prevents them from fraying enough to kill a cell — speculation ran wild about its role in aging and cancer, setting off a full-court press to produce drugs to activate or block the enzyme.
While neither telomerase-based anti-aging drugs, touted as a “fountain of youth,” nor anticancer drugs have yet appeared, the publication today by UC Berkeley scientists of the first detailed picture of the molecular structure of human telomerase should jump-start that effort, allowing more targeted drug screens and intelligent design of new drugs.
“It has been a long time coming. It took a lot of persistence,” said Kathleen Collins, a UC Berkeley professor of molecular and cell biology who has worked on the enzyme for 26 years.
Collins and Eva Nogales, also a professor of molecular and cell biology, are the senior authors of a paper describing the 3-D molecular structure of the human telomerase enzyme published this week in the journal Nature.
One bottleneck has been obtaining pure samples of this complex molecule, which is composed of an RNA backbone decorated by six types of protein that move around as they add DNA to the ends of chromosomes. Labs around the world have debated whether the enzyme operates singly or as conjoined twins, and how and how many proteins decorate the RNA backbone.
Without consensus on these questions, it has proven difficult to design a drug to target the molecular machine and either destroy telomerase activity – which could stop a cancer that has boosted its telomerase levels – or restart telomerase, perhaps to prime the body for rapid cell division after a bone marrow transplant.
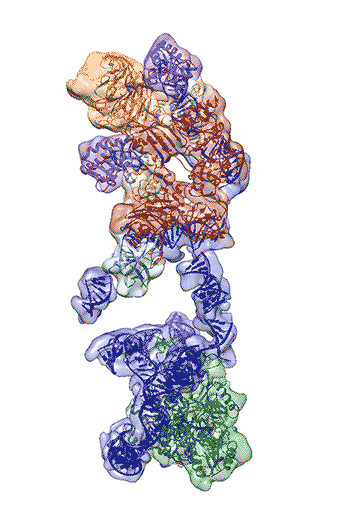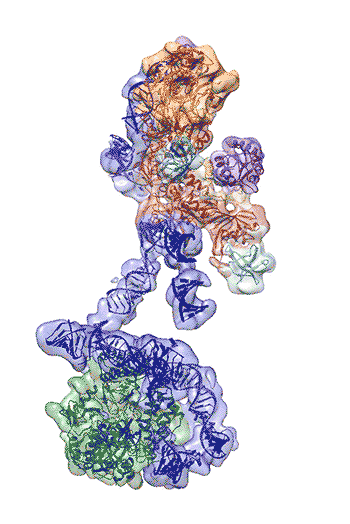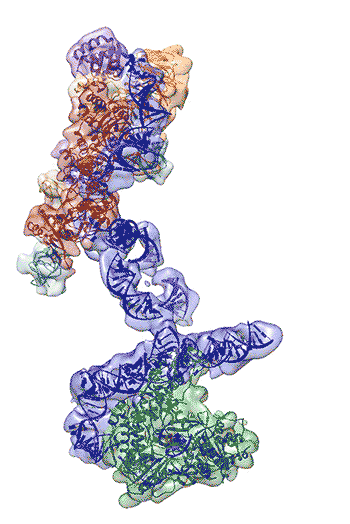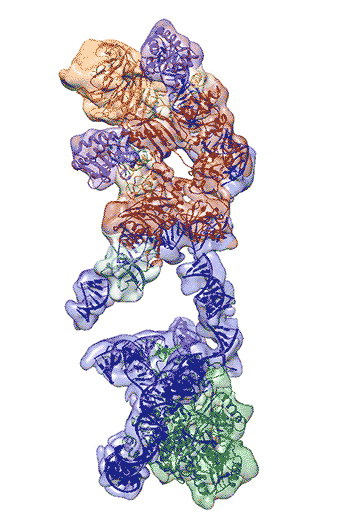
The newly revealed structure still lacks fine detail, but combined with knowledge of the gene sequence of human telomerase, it provides enough information to start thinking about potential targets for drugs, said first author Thi Hoang Duong “Kelly” Nguyen, a Miller Institute postdoctoral fellow at UC Berkeley.
“The best previous images of human telomerase had a resolution of only 30 Ångstroms; we were able to get about 7 to 8 Ångstroms resolution using cryoelectron microscopy,” Kelly said. “When I got to the point where I could see all the subunits – we had 11 protein subunits in total – it was a moment of, ‘Wow, wow, this is how they all fit together.’”
Nguyen, Collins and Nogales are actively working to improve the resolution to 3 or 4 Ångstroms – about the size of two carbon atoms – which is sufficient for drug design.
Telomerase tops up the telomeres
Telomeres were first detected at a molecular level in the late 1970s by Elizabeth Blackburn, then at UC Berkeley and now president emerita of the Salk Institute for Biological Studies in La Jolla, California. Working with the ciliated protozoan Tetrahymena, she and colleagues showed that the ends of the chromosomes are capped by repeating sequences of DNA. Armed with knowledge of telomere sequence, researchers then showed that telomeres in tissues of multicellular organisms grow shorter each time a cell divides. The telomeres protect the DNA strands from fraying and getting damaged at their ends, much like the plastic tip on the end of a shoelace. The fact that they drop off with each cell division is thought to protect us from cancer, when a cell is hijacked and proliferates continually.
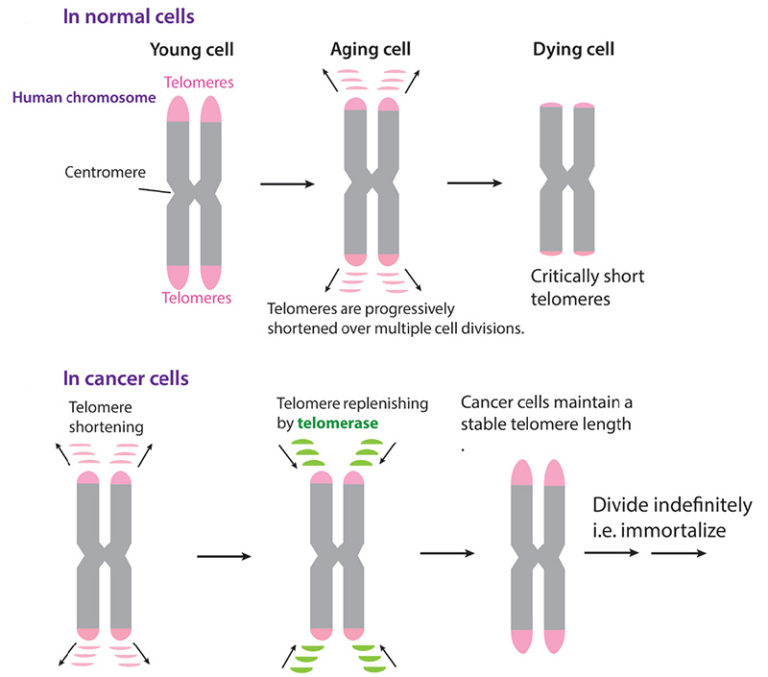
In 1985 at UC Berkeley, Blackburn and then-graduate student Carol Greider discovered telomerase, an enzyme that adds DNA back to the ends of chromosomes, lengthening them and extending the lifespan of the cell. Blackburn, Greider and another colleague, Jack Szosak, shared the 2009 Nobel Prize in Physiology or Medicine for the discovery of telomerase.
Scientists have since found that, in humans and other multicellular organisms, telomerase is expressed only in the embryo, not in most adult cells. That means that most cells at birth have a predetermined ability to grow and divide, after which they die. Many scientists believe that depleted telomeres are a major cause of aging.
Collins has been trying to determine the structure of telomerase ever since the first human telomerase protein was discovered in 1997, and she and her colleagues have discovered and extensively characterized many of the proteins in the large enzyme, as well as the broken-up hairpin structure of the RNA backbone of telomerase. Yet how they all fit together was an enigma because of conflicting results from many different labs.
Nguyen was able to isolate the active enzyme and purify it much better than anyone had before, and employed a new, state-of-the-art cryoelectron microscope to determine the structure of the active telomerase unambiguously. Cryo-EM is a technique for determining molecular structures of compounds that cannot be crystallized and imaged with X-rays, and its developers won the 2017 Nobel Prize in Chemistry.
According to Nogales, cryo-EM is ideally suited to studying human telomerase because it can deal with the extremely small amounts of purified sample, and it can produce a structure even when the molecule under study is highly flexible, as is the case with human telomerase.
Once the UC Berkeley team had the structure, Nguyen said, it became clear why genetic mutations in some of the proteins in telomerase interfere with the enzyme to cause disease. In 1999, Collins discovered the first known human disease caused by a telomerase mutation: a mutation in a telomerase protein called dyskerin that is responsible for a rare disease called dyskeratosis congenita. Patients develop anemia as well as skin and gut problems, and most frequently die from bone marrow failure.
The reason, Collins says, is that there are two dyskerin molecules bound to the RNA backbone that have to not only reach out to the network of other proteins but also touch one another, and disease-causing mutations prevent these linkages, crippling the ability of the RNA backbone to survive in cells. Some children with dyskeratosis congenita have telomerase levels about 25 percent of normal and a lifespan of less than two decades. Those with half the normal level of telomerase typically reach a health crisis in mid-life.
Collins is ecstatic to finally have a definitive structure for telomerase and looks forward to learning more about the intricate assembly process of one of the most complex enzymes in the body: a polymerase as complicated as the ribosome, which reads RNA to produce proteins.
“I didn’t think it would be this complicated when I decided to study this molecule,” she said. “I became curious about how telomerase works in 1991, when I was just finishing as a grad student, and I was looking for a really simple polymerase system in which to understand nucleic acid interactions. And I thought, my god you can’t be simpler than this. That was super naive.”
The work was supported by the National Institutes of Health (GM054198) and the Miller Institute for Basic Research. Other UC Berkeley co-authors are Jane Tam, Robert Wu, Basil Greber and Daniel Toso. Nogales is a Howard Hughes Medical Institute investigator, and with Collins a member of the California Institute for Quantitative Biology.
