Genome sequenced for amoeba that flips into free-swimming cell
In the long evolutionary road from bacteria to humans, a major milestone occurred some 1.5 billion years ago when microbes started building closets for all their stuff, storing DNA inside a nucleus, for example, or cramming all the energy machinery inside mitochondria.

Scientists have now sequenced the genome of a weird, single-celled organism called Naegleria gruberi that is telling biologists about that transition from prokaryotes, which function just fine with all their proteins floating around in a soup, to eukaryotes, which neatly compartmentalize those proteins.
The sequence, produced by the Department of Energy Joint Genome Institute (JGI), and an analysis by scientists from the University of California, Berkeley, Lancaster University in the United Kingdom and institutions elsewhere in the United States and the U.K. are published in the March 5 issue of the journal Cell.
"In a sense, analyzing the Naegleria genome shows us what it would be like to be on this planet more than a billion years ago, and what kind of organisms were around then and what they might have looked like," said Simon E. Prochnik, a JGI and UC Berkeley bioinformaticist and coauthor of the Cell paper.
Naegleria is a common soil amoeba - the sequenced organism was isolated from the mud in a grove of eucalyptus trees on the UC Berkeley campus - that, under stress, quickly grows two flagella, like sperm tails, that it uses to swim around. It has a third identity, a hard cyst, that can persist in the soil until conditions become damp and warm enough for it to turn into an amoeba.
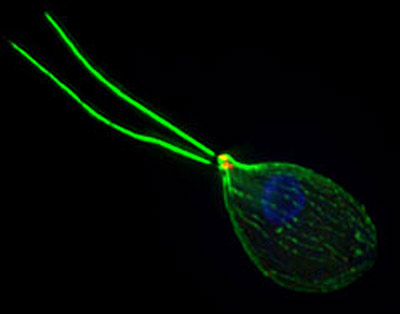
"This one-celled organism hunts and eats bacteria as an amoeba, swims around looking for a better environment as a flagellate, and then hunkers down and waits for good times as a cyst," Prochnik said. "It is a very rare process to go from amoeba to flagellate like this."
Not surprisingly, the organism is packed with genes that help support these three personalities, he said. He and his colleagues report that this amoeboflagellate contains 15,727 genes coding for proteins, while humans have 23,000 protein-coding genes.
"Naegleria has a lot of genes because it has a complicated lifestyle; most single-celled organisms - in particular, parasites - have a simpler lifestyle, and therefore have fewer genes," Prochnik said. "These single-celled organisms are highly versatile, containing all the genetic information necessary to survive in a wide range of environments and under a wide range of stresses."
The researchers compared the Naegleria genome to the genomes of 16 other eukaryotes, ranging from humans and fungi to green plants and other unicellular eukaryotes, shedding light on the set of perhaps 4,000 genes that may have been part of the first, most primitive eukaryotes, according to UC Berkeley graduate student Lillian Fritz-Laylin, first author of the paper. The number of genes surprised the researchers, because previous genome comparisons that included parasites came up with a much lower number. That may be because parasites live off their host and have been able to shed many genes that are critical for a free-living organism, they said.
"Now that our analysis focuses on data from free-living organisms, including Naegleria, that haven't lost all these genes and functions, we can make a broader comparison, and we find a lot more proteins were probably present in the eukaryotic ancestor than we previously thought," Fritz-Laylin said.
"This is the first genome comparison that includes not only Naegleria, but representatives of all six sequenced groups of eukaryotes," Prochnik said. Naegleria is part of a diverse group that includes a cousin, Naegleria fowleri, that can fatally infect swimmers. The other eurkaryotic groups are animals and fungi; plants and green algae; chromalveolata, which include diatoms, red tide and malaria; amoebozoa, which include various single-celled amoebae; and the diverse group that includes parasites like giardia.
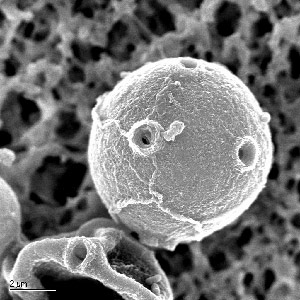
Among other things, Naegleria's genes shed light on how cells move, how they signal one another and how they metabolize nutrients.
As an amoeba, Naegleria pushes out little feet, called pseudopods, that propel it in its hunt for food. Yet, once the food disappears, the amoeba creates flagella from scratch and uses them to swim about in search of new hunting grounds.
What is interesting, Fritz-Laylin said, is that pseudopods and flagella use different proteins for movement. Amoebae make use of actin, which provides the internal scaffolding for the cell and for the pseudopods that help amoebae explore their environment. Flagella, on the other hand, are made mostly of the protein tubulin. Because Naegleria has both types of movement, the organism can help scientists understand the origins of these parallel systems during the evolution of eukaryotes.
Scientists can starve populations of Naegleria in its amoeba form and have seen it switch quickly and simultaneously to its flagellar form. This suggests that the switch from an actin-based system to a microtubule-based system of movement is very highly regulated and synchronized across a population.
"The sequence helped us identify the genes associated with each type of motility," she said. "Although this has been done for flagellar motility, it had not been done for amoeboid motility."
The genome also reveals versatility in how Naegleria produces energy. The organism can use oxygen to burn nutrients - glucose, amino acids or fatty acids - for energy or, in the absence of oxygen, utilize other nutrients and possibly produce hydrogen as a byproduct.
Like the recently sequenced, free-living alga Chlamydomonas, Naegleria likely uses its metabolic flexibility to survive the intermittent hypoxia common to muddy environments, the researchers concluded. Prochnik suggests that Naegleria could help biologists understand hydrogen production that, in other organisms, might be used to produce energy.
Fritz-Laylin noted that, while the genome will be a boon to the small number of biologists who study the organism, it also will help in understanding the evolution of more complicated organisms.
"By comparing diverse organisms like Naegleria from all over the family tree of eukaryotes we can begin to understand where we come from," she said.
Other co-authors of the paper are Michael L. Ginger of the School of Health and Medicine at Lancaster University; Meredith L. Carpenter, Alex Paredez, W. Zacheus Cande and Daniel S. Rokhsar of UC Berkeley; Rochak Neupane of UC Berkeley's Center for Integrative Genomics; Alan Kuo, Jarrod Chapman, Shengqiang Shu, Asaf Salamov, Erika Lindquist, Hank Tu, Harris Shapiro, Susan Lucas and Igor V. Grigoriev of JGI; Joel B. Dacks of the University of Alberta Edmonton in Alberta, Canada; Jonathan Pham, Michael Cipriano and Scott C. Dawson of UC Davis; Joel Mancuso of Gatan Inc. in Pleasanton, Calif.; Mark C. Field of the University of Cambridge, U.K.; and Chandler Fulton of Brandeis University in Waltham, Mass. Cande, Dawson, Fulton and Rokhsar were co-principal investigators for the seqeuncing project. Rokhsar is the program head for computational genomics at JGI and a professor of molecular and cell biology and of physics at UC Berkeley.
Funding for the project came primarily from the Department of Energy.
