Could Liposomes Be the Unsung Heroes of the Pandemic?
Liposomes may be the unsung heroes of the COVID-19 pandemic. Without the protection of these microscopic vesicles, the delicate strands of messenger RNA (mRNA) that lie at the heart of the Pfizer and Moderna COVID-19 vaccines would be quickly destroyed by enzymes in the body, making it nearly impossible for their genetic instructions to reach the insides of human cells.
But vaccine delivery isn’t the only way that these particles can be used in the battle against COVID-19. In a new study, a team of engineers at the University of California, Berkeley, attached SARS-CoV-2 “spike” proteins to the surface of liposomes, creating lab-made mimics of the deadly virus which the researchers call “spike-liposomes.” These spike-lipsomes can be used to test the efficacy of neutralizing antibodies that could potentially be used to treat COVID-19 patients.
The study also demonstrates how a new DNA-patterning technique, developed by the team last year, can help scientists rapidly characterize and conduct experiments on a variety of different types of liposomes, and their cousins, lipid nanoparticles.
“Lipid nanoparticles are really relevant to a number of biomedical applications: They’ve been used in drug delivery for decades, and they can also serve as models of viruses that have membranes on the outside, including coronaviruses,” said study lead author Molly Kozminsky, a postdoctoral fellow in the Sohn Research Lab at UC Berkeley. “We actually developed these spike-liposomes because we wanted to test a new COVID-19 diagnostic method that we were developing in the lab. But first, we needed a way to validate that these particles were displaying the SARS-CoV-2 spike protein correctly, and we realized that our DNA-patterning technique would allow us to do this and other exciting experiments in a very efficient way.”
Liposomes are tiny, spherical vessels constructed of lipid membranes very similar to those that encase most biological cells. And, in the same way that the membranes of biological cells are dotted with a variety of proteins that help the cell interact with the outside world, researchers have learned to attach different types of proteins into the membranes of liposomes, giving the particles different functions and abilities.
Excitement over liposomes has been most pronounced in the pharmaceutical industry, where drugmakers have experimented with outfitting liposomes with proteins that only interact with very specific cells in the body, allowing them to target the delivery of drug molecules to only the tissues where they are needed. As Kozminsky points out, liposomes can also be used to create simple models of viruses and other pathogens that have lipid membranes, including SARS-CoV-2.
However, researchers must first verify that the liposome proteins are able to properly interact with their environment. For example, the SARS-CoV-2 spike protein binds with proteins on human cells called the ACE2 receptors, triggering a series of events that lets the virus fuse with the cell.
“For the spike-liposomes, we wanted to make sure that the spike protein that we put on the surface of the liposome was in the correct configuration to allow it to bind to ACE2 receptors,” Kozminsky said. “If that was the case, then the way that these spike-liposomes are formulated would likely also model the way that the SARS-CoV-2 virus spike protein interacts with cells, antibodies and other proteins”
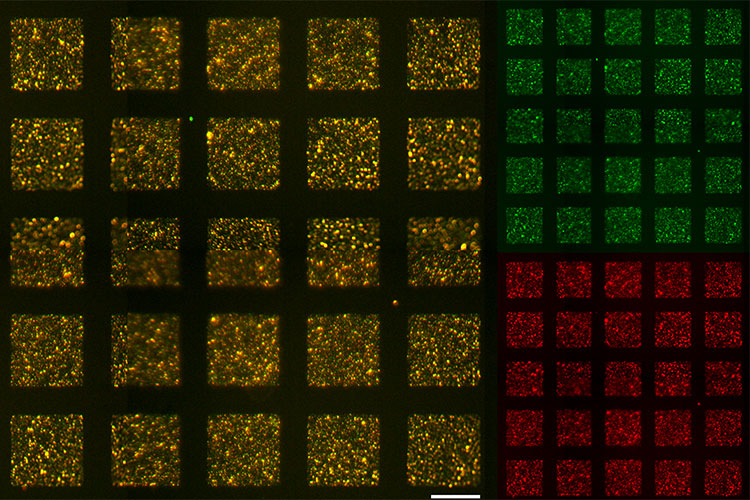
Kozminsky realized that the DNA-printing technique, which was originally developed by the Sohn Lab to “print” different types of cells into patterns that model biological tissues, could also be used to quickly verify that the spike-liposomes were presenting the SARS-CoV-2 spike protein correctly.
“We knew we had to test the liposomes first, and when we looked at all the ways we’d have to validate the liposomes, we found that the techniques were somewhat arduous,” Kozminsky said. “We realized how much easier it would be to use our DNA-directed printing technology.”
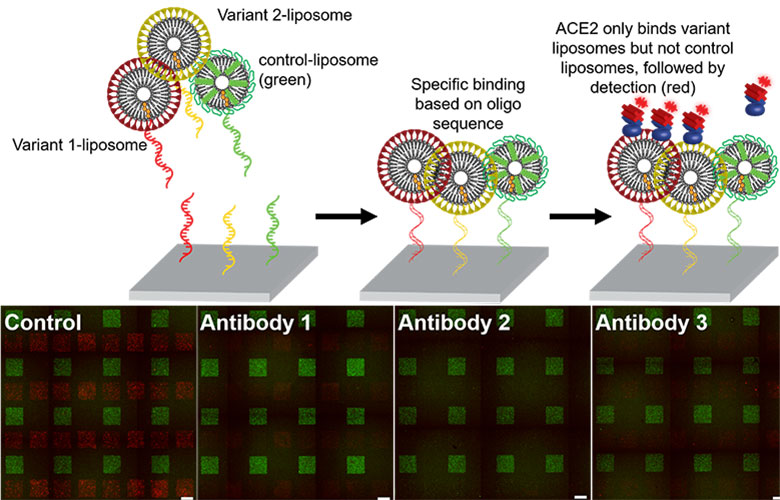
To conduct the experiment, Kozminsky printed the spike-liposomes onto a slide, and then tagged them with a green fluorescent protein. She then washed the slide with ACE2 receptor proteins that had been tagged with a red fluorescent protein. When she imaged the slide, she found that most of it glowed red, indicating that the ACE2 receptor proteins were binding to the spike-liposomes on the slide. Kozminsky then repeated the experiment with cells that express the ACE2 receptor, showing that they, too, were able to bind with the spike-liposomes.
To show how the spike-liposomes could be used to test the effectiveness of COVID-19 treatments, Kozminsky created two different types of spike-liposomes, each displaying a different variant of the SARS-CoV-2 spike protein. After using DNA-printing to pattern these onto microscope slides, she washed the slides with three different types of commercially available neutralizing antibodies against variants of the SARS-CoV-2 spike protein. She then tested whether the presence of these neutralizing antibodies successfully prevented the ACE2 receptor proteins from binding to the spike-liposomes, and found that the results were consistent with those reported by the antibody manufacturers.
“What’s really cool about this technique is that it is very high-throughput, meaning that you can run experiments using lots of different combinations of liposomes at once,” said study senior author Lydia Sohn, the Almy C. Maynard and Agnes Offield Maynard Chair in Mechanical Engineering at UC Berkeley. “So, for example, drug companies could use this technique to very quickly test which antibodies will work most effectively against a particular variant of SARS-CoV-2. Or, it could be used to screen new proteins for targeted drug delivery, to make sure that that protein targets particular cell types in the body. It really adds a new strategy for combating this virus.”
Thomas R. Carey of the UC Berkeley-UC San Francisco Graduate Program in Bioengineering is a co-author of this study. This work was supported in part by CITRIS COVID-19 Response Seed Funding, by the National Institutes of Health National Cancer Institute (grant #1F32CA243354-01), by the University of California Cancer Research Coordinating Committee Fellowship and by a generous gift from Michael and Margaret Checca.
RELATED INFORMATION
