Computer Simulations Yield Clues to How Cells Interact With Surroundings
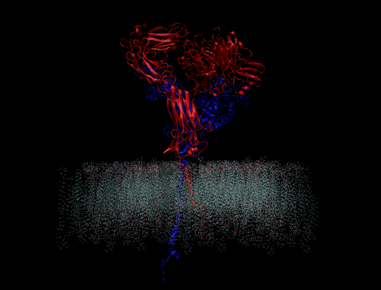
Your cells are social butterflies. They constantly interact with their surroundings, taking in cues on when to divide and where to anchor themselves, among other critical tasks.
This networking is driven in part by proteins called integrin, which reside in a cell’s outer plasma membrane. Their job is to convert mechanical forces from outside the cell into internal chemical signals that tell the cell what to do. That is, when they work properly. When they misfire, integrins can cause diseases such as atherosclerosis and several types of cancer.
Despite their importance—good and bad—scientists don’t exactly know how integrins work. That’s because it’s very difficult to experimentally observe the protein’s molecular machinery in action. Scientists have yet to obtain the entire crystal structure of integrin within the plasma membrane, which is a go-to way to study a protein’s function. Roadblocks like this have ensured that integrins remain a puzzle despite years of research.
But what if there was another way to study integrin? One that doesn’t rely on experimental methods? Now there is, thanks to a computer model of integrin developed by Berkeley Lab researchers. Like its biological counterpart, the virtual integrin snippet is about twenty nanometers long. It also responds to changes in energy and other stimuli just as integrins do in real life. The result is a new way to explore how the protein connects a cell’s inner and outer environments.
“We can now run computer simulations that reveal how integrins in the plasma membrane translate external mechanical cues to chemical signals within the cell,” says Mohammad Mofrad, a faculty scientist in Berkeley Lab’s Physical Biosciences Division and associate professor of Bioengineering and Mechanical Engineering at UC Berkeley. He conducted the research with his graduate student Mehrdad Mehrbod.
They report their research in a recent issue of PLoS Computational Biology.
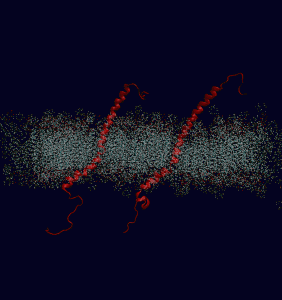
Their “molecular dynamics” model is the latest example of computational biology, in which scientists use computers to analyze biological phenomena for insights that may not be available via experiment. As you’d expect from a model that accounts for the activities of half a million atoms at once, the integrin model takes a lot of computing horsepower to pull off. Some of its simulations require 48 hours of run time on 600 parallel processors at the U.S. Department of Energy’s (DOE) National Energy Research Scientific Computing Center (NERSC), which is located at Berkeley Lab.
The model is already shedding light on what makes integrin tick, such as how they “know” to respond to more force with greater numbers. When activated by an external force, integrins cluster together on a cell’s surface and join other proteins to form structures called focal adhesions. These adhesions recruit more integrins when they’re subjected to higher forces. As the model indicates, this ability to pull in more integrins on demand may be due to the fact that a subunit of integrin is connected to actin filaments, which form a cell’s skeleton.
“We found that if actin filaments sustain more forces, they automatically bring more integrins together, forming a larger cluster,” says Mehrbod.
The model may also help answer a longstanding question: Do integrins interact with each other immediately after they’re activated? Or do they not interact with each other at all, even as they cluster together?
To find out, the scientists ran simulations that explored whether it’s physically possible for integrins to interact when they’re embedded in the plasma membrane. They found that interactions are likely to occur only between one compartment of integrin called the β-subunit.
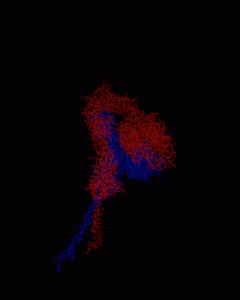
They also discovered an interesting pattern in which integrins fluctuate. Two integrin sections, one that spans the cell membrane and one that protrudes from the cell, are connected by a hinge-like region. This hinge swings about when the protein is forced to vibrate as a result of frequent kicks from other molecules around it, such as water molecules, lipids, and ions.
These computationally obtained insights could guide new experiments designed to uncover how integrins do their job.
“Our research sets up an avenue for future studies by offering a hypothesis that relates integrin activation and clustering,” says Mofrad.
The research was supported by a National Science Foundation CAREER award to Mofrad. NERSC is supported by DOE’s Office of Science.
