Bottlenecks That Reduced Genetic Diversity Were Common Throughout Human History
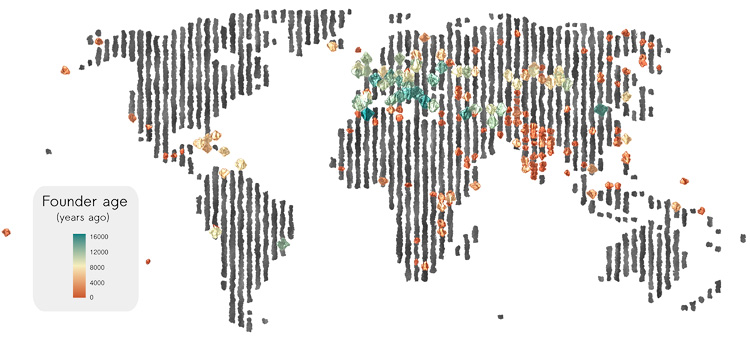
Human populations have waxed and waned over the millennia, with some cultures exploding and migrating to new areas or new continents, others dropping to such low numbers that their genetic diversity plummeted. In some small populations, inbreeding causes once rare genetic diseases to become common, despite their deleterious effects.
A new analysis of more than 4,000 ancient and contemporary human genomes shows how common such “founder events” were in our history. A founder event is when a small number of ancestral individuals gives rise to a large fraction of the population, often because war, famine or disease drastically reduced the population, but also because of geographic isolation — on islands, for example — or cultural practices, as among Ashkenazi Jews or the Amish.
More than half of the 460 groups represented by these individuals had experienced a population bottleneck somewhere in their past that decreased their genetic diversity and likely increased the incidence of recessive hereditary diseases.
The analysis by population geneticists at the University of California, Berkeley, is the first comprehensive look at founder events across a broad swath of human populations over the past 10,000 years or so of human history and pinpoints when these events occurred.
According to the authors, the findings will be useful not only to archeologists and historians tracking the movement and mixing of populations around the world, but also to scientists and doctors studying human genetic variation. The genetic diseases of inbred populations have helped scientists find many disease-causing mutations in the human genome and discover the causes of numerous genetic and inherited diseases.
“Genomic data is really powerful because it not only tells us about where we come from, it tells us about our history at various different time scales, and you can look at how closely related different individuals are to each other,” said senior author Priya Moorjani, an assistant professor of molecular and cell biology at UC Berkeley. “But also, it tells us about bits of DNA that are functionally important and can cause diseases. So, they become quite important to study from a biomedical perspective.”
Many of the populations represented by individuals in the sample were or are much more inbred than ethnic Ashkenazi Jews, who some scientists have estimated once dwindled to a population of less than a couple of thousand individuals about 1,000 years ago. The Onge, a group in the Andaman Islands of the Indian Ocean, underwent a population bottleneck 10 times more extreme than that of Ashkenazi Jews, and today it numbers only about 100 individuals.
The researchers found that many Native American populations and groups from Oceania and South Asia also suffered severe population bottlenecks. Some coincide with known historical events — for instance, the residents of Rapa Nui (Easter Island) underwent a founder event about 260 years ago, coincident with the migration of Europeans to the island.
Others correlate well with the known movement of peoples into an area and with changing cultural artifacts and practices. For example, Anatolian farmers and Eurasian steppe pastoralists moved into Europe between about 4,000 and 10,000 years ago, and the groups intermingled with existing European hunter-gatherers.
“The first surprise was that over half the groups we surveyed had evidence for founder events,” Moorjani said. “So, it’s not just Ashkenazi Jews or Finns that have a unique history, but many populations living today have had strong founder events — in fact, stronger founder events than these two groups, like several contemporary South Asian groups, hunter-gatherers or populations living on islands. And many of these groups would be really important for prioritizing functional studies. We have learned so much about genetic variation from groups like Ashkenazi Jews and Finns that the potential for discovery is really high if we can expand these studies to other worldwide populations.”
Moorjani, former UC Berkeley undergraduate Gillian Chu and first author Rémi Tournebize, now a postdoctoral fellow at the Instituto Gulbenkian de Ciência in Oeiras, Portugal, published their findings today (June 23) in the journal PLOS Genetics.
Working with incomplete ancient DNA
The study was made possible by a genomics analysis program called ASCEND (Allele Sharing Correlation for the Estimation of Non-equilibrium Demography), which was created by Tournebize and Moorjani specifically to analyze partial genome sequences — in particular, ancient DNA. This DNA is generally sequenced from bones or teeth that are hundreds to thousands of years old and represent not only our Homo sapiens ancestors, but other human groups, like Neanderthals and Denisovans.
Such DNA is typically damaged so that only a portion of the individual’s genome can be sequenced. But since human genomes contain about 3 billion base pairs of DNA, even a mere 100,000 base pairs can provide information about that person’s heritage, Moorjani said. Many genome analysis programs today work only with nearly complete genome sequences, primarily from contemporaneous peoples.
“While ancient DNA is really powerful, one of the challenges is that it has much lower quality compared to data from living people, because once an individual dies, the DNA starts degrading, and it’s very hard to recover very high quality data compared to present-day individuals,” Moorjani said. “But the majority of the demographic inference methods are built thinking that you can get large numbers of samples from populations and high-quality data across the genome. Our methods were developed to leverage this low-coverage, highly degraded DNA to really understand our evolutionary history.”
ASCEND measures the sharing of DNA between individuals within and across populations. When a population undergoes a founder event, its size dwindles to a few individuals. The offspring of these founder individuals, in turn, share long blocks in their genome that are inherited “identical by descent” from these few ancestors. As time passes, these blocks will become smaller due to crossover events that occur during meiosis, when chromosomes duplicate and mix before segregating into sperm or egg cells. The rate of crossovers is well characterized and provides a kind of molecular clock. The ASCEND program compares how large the shared blocks are within individuals in a population to infer when the individuals might have shared a common ancestor, that is, when a founder event occurred in the population’s history. A large-scale, pair-wise statistical comparison of genomic DNA allows an estimation of when and how intense the bottleneck was.
The genome data came from the Allen Ancient DNA Resource, a database created by David Reich and collaborators at Harvard University, with whom Moorjani earned her Ph.D. The public database currently includes available present-day and ancient genomes from more than 14,000 individuals and more than a million common mutations or variants — single nucleotide polymorphisms, or SNPs — within those DNA sequences. At the time Moorjani started her study, the database held fewer ancient and modern genomes. She and Tournebize focused on the genomes of 2,310 present-day individuals from 184 groups, then expanded their study to look at an additional 1,947 individuals representing 164 worldwide ancient populations.
“Applying this method, we uncovered founder events that had not been identified previously, for instance, in populations from ancient Morocco or Siberia,” Tournebize said. “As a French guy, I was really surprised to discover a founder event in Basque people, dated around the 1st century BCE and possibly related to Roman colonization of this region. We’ll need more genetic data, especially from ancient samples, and collaboration with social scientists to understand the detailed historical events that might be associated with this bottleneck.”
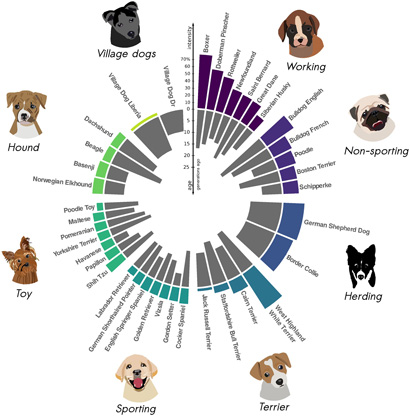
To test the ASCEND program in other species, Moorjani and Tournebize turned to dogs. The genome sequences of about 40 modern dog breeds are available, so the researchers ran them through the program to determine how long ago founder events occurred in breeds ranging from African village dogs — the least inbred — to breeds like boxers, dobermans and rottweilers, the most inbred. Consistent with the establishment of many dog breeds during Victorian times, they confirmed extreme founder events in most breeds within the last 25 generations, that is, 75 to 125 years.
“Dogs are so interesting that it was exciting to expand the analysis to another species, but it was really sad to see how strong the founder events are,” she said. “Most dogs these days have so many more problems than village dogs. Their rates of cancers and congenital diseases are pretty high. And that’s largely because of these very severe founder events in their history during breed formation.”
Population mixing
In another recent paper, Moorjani and her colleagues described a different genomics analysis program that analyzes a single individual’s genome, whether complete or partial, and estimates the amount of admixture of other populations over time. The researchers used this program, called DATES (Distribution of Ancestry Tracts of Evolutionary Signals), to analyze about 1,100 ancient genomes and reconstruct major gene flow events in Europe since about 10,000 BCE.
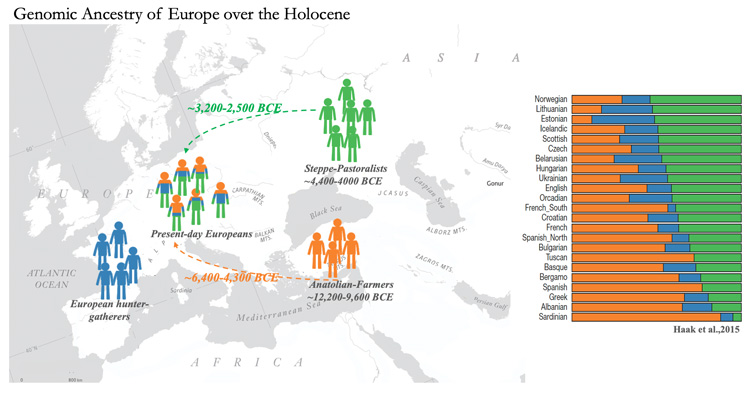
One surprising finding was that the genomes of Anatolian farmers, who lived in what is today Turkey, show admixture of genes from Iranian Neolithic farmers long before the advent of agriculture in Anatolia. This suggests that farming did not originate in Anatolia, as many archeologists have suggested.
“We had samples of Anatolian hunter-gatherers who don’t have Iranian ancestry and samples of Anatolian early farmers who have Iranian ancestry, but we didn’t know when this mixture occurred,” she said. “In our case, we were able to actually figure out the key time point when this group formed, which predates agriculture in the region. And based on that, we are able to tell that farming must have spread through cultural diffusion, rather than having originated in Anatolia.”
Another discovery was the timing for the formation of Bronze Age steppe pastoralists. These groups made a large impact, both genetically and demographically, in Eurasia during the Bronze Age and, according to some studies, are responsible for the spread of Indo-European languages. Archeological studies suggest these groups inhabited regions of the steppe in present day Russia and Ukraine from 3,300 to 2,600 BCE. Using the genetic dating method, the researchers found these groups were genetically formed between 4,400 and 4,000 BCE, predating previous findings by over a half a millennium.
“Our study emphasizes the power of dating population mixtures and formation, rather than just using temporal sampling and tracking the presence or absence of a particular ancestry in ancient samples, which is highly dependent on sampling choice and density,” said UC Berkeley postdoctoral fellow Manjusha Chintalapati, first author of the paper.
Moorjani plans to use ASCEND and DATES to take a closer look at many ancient populations, in particular those in India, that have strong founder events that suggest the possibility of many unrecognized recessive diseases that could help to reduce disease burden in the group and shed light on the basic functions of human genes.
“In our analysis, we find that 64% of South Asian populations have very strong founder events, so we are trying to do targeted sample collection in these groups to characterize some of the deleterious variants due to the founder events,” she said.
DATES, for example, suggests that each isolated population in South Asia has admixtures of local indigenous hunter-gatherers, Near Eastern farmers and Steppe pastoralists or herders, but in different proportions that remained the same for many hundreds of generations. Strikingly, most European populations also derive ancestry from similar three groups, though the groups have continued to freely mix with each other after the initial mixture.
“It’s really exciting to do this work at Berkeley, where Allan Wilson’s lab came up with the idea of a molecular clock, and to continue on his path to use genomic data for learning about the timing of different evolutionary events,” Moorjani said, referring to the late biochemist and pioneer of molecular evolution, who died in 1991.
The two studies were funded by the Burroughs Wellcome Fund, a Sloan Research Fellowship and the National Institutes of Health (R35GM142978).
