Biologists home in on paleo gut for clues to our evolutionary history
For all the anxiety today about the bacteria in our gut being under constant assault by antibiotics, stress and bad diets, it turns out that a lot of the bacteria in our intestines have been with us for at least 15 million years, since we were pre-human apes.

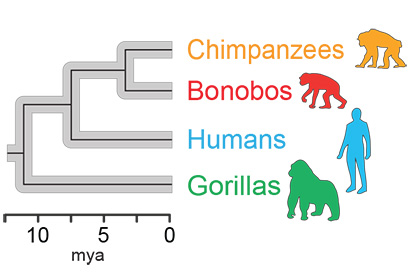
A new comparison of the gut microbiomes of humans, chimps (our closest ancestor), bonobos and gorillas shows that the evolution of two of the major families of bacteria in these apes’ guts exactly parallels the evolution of their hosts.
This shows that the microbes in our guts are determined in part by our evolutionary history, not just external factors like diet, medicine and geography. Obesity, cancer and some inflammatory diseases, such as diabetes and Crohn’s disease, have been linked to imbalances in the mix of microbes in our stomach and intestines.
“We are showing that some human gut bacteria are the direct descendents of gut bacteria that lived within our common ancestors with apes,” said lead researcher Andrew Moeller, a Miller Postdoctoral Fellow in UC Berkeley’s Museum of Vertebrate Zoology. “It shows there has been an unbroken line of inheritance or transfer from one generation to another for millions of years, since the dawn of African apes.”
Moeller is beginning to assemble a snapshot of the microbes in the guts of our ancient ape ancestor — in essence, a paleo gut that fit our paleo diet — and hopes to go even further back in time if, as seems likely, all mammals have evolved their unique microbiota from a common ancestral population in the distant past.
“We now have samples from all the major groups of mammals, and we’re working on tracing the evolution of the microbiome all the way back to when we were tiny carnivorous creatures 100 million years ago,“ he said.
Moeller and his African and American colleagues, including corresponding author Howard Ochman at the University of Texas at Austin, will publish their findings in the July 22 issue of Science.
“It’s surprising that our gut microbes, which we could get from many sources in the environment, have actually been co-evolving inside us for such a long time,” said Ochman, who noted that the microbes were passed down over hundreds of thousands of host generations.
Microbes are us
The importance of the human microbiome — the microbes that live on our skin, in our bodily orifices and in our intestines — has become evident over the past decade as scientists have sequenced the microbial populations from healthy and unhealthy people, from newborns to seniors, and of peoples around the world. There are about as many microbial cells in our body as our own cells, and they appear to be transmitted from mother to child during birth and continually altered by the home environment, type of diet, medications and even the family pet. Gut bacteria, in particular, guide the early development of our intestines, train our immune systems to fight pathogens and may even affect our moods and behavior.
“We know that gut bacteria are really integrated with our biology, and this reseach gives us a framework for investigating how that has come to be,“ Moeller said.
Most researchers studying the gut microbiota of humans or animals have identified the specific microbes present based on a distinctive bit of DNA that codes for a piece of RNA in the bacterial ribosome, which manufactures proteins. This piece of DNA, called the 16S subunit of ribosomal RNA, does not change quickly over time, however, so it gives a very coarse timeline for the evolution of bacteria. Its slow rate of change does not allow researchers to determine which individual bacterial strains — different variants of the same species — are present, only the genus and sometimes species.
Moeller and his colleagues decided, instead, to look at a more rapidly evolving gene that varies even in different strains of bacteria, allowing them to identify individual bacterial strains. The gene, called gyrase B, codes for a variable subunit of the DNA gyrase protein, which helps DNA coil and uncoil.
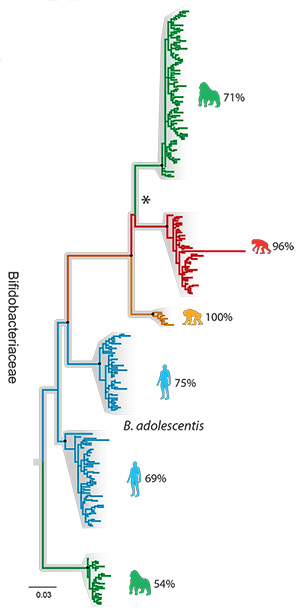
He and his colleagues obtained feces from 24 gorillas living in Cameroon, 47 chimpanzees from Gombe National Park in Tanzania, 24 wild bonobos from the Democratic Republic of the Congo and 16 people from Connecticut. They isolated bacterial DNA from these samples, amplified the DNA of the gyrase B genes present and catalogued the variations within three major bacterial families to create a microbiome family tree for three families of bacteria — Bacteroidaceae, Bifidobacteriaceae and Lachnospiraceae — that together comprise about 20 percent of all the microbes in the human gut, including both bacteria and their cousins, the Archaea.
Only bacteria in the Bacteroidaceae and Bifidobacteriaceae families showed cospeciation with their ape hosts, with gut microbial diversity lowest in humans and highest in gorillas.
“Once we calibrated the molecular clock, we were able to date the split of human and chimp bacteria at around 5.3 million years ago, and the human-gorilla gut bacteria split at around 15.6 million years ago, which are roughly in line with what we know from fossil and genomic data of the hosts,“ Moeller said. “It is one more line of evidence that gut bacteria have cospeciated with humans.“
“In a way, host speciation is like continental drift: When two continents drift apart, whole biotas begin to diverge,” Moeller said. “Here, as the hosts are splitting, a good chunk of their microbiota is also splitting and diversifying.”
The third bacterial family tree, the Lachnospiraceae, was more complicated. There were apparently at least four times when these bacteria were transferred between different host species. The researchers speculate that because these bacteria form spores and can thus survive outside their hosts for long periods, they were easily passed between species.
“We have shown that the microbiome is a composite of microbial lineages, some that have cospeciated with us, and some that have been passed around from one host species to another,“ he said.
They also looked at the microbial metagenomes of Africans from Malawi, and saw distinct differences between the African and the U.S. human microbiota, with some lineages seeminly lost by Americans.
“It will be interesting in the future to do a full study of human populations using this strain-level method to see whether we can use bacteria to reconstruct the history of human migrations,“ he said.
Other coauthors are Alejandro Caro-Quintero at Corpoicá C.I Tibaitata (Colombia); Deus Mjungu at the Gombe Stream Research Center (Tanzania); Alexander Georgiev at Northwestern University and Harvard University; Elizabeth Lonsdorf at Franklin & Marshall College; Martin Muller at the University of New Mexico; Anne Pusey at Duke University; Martine Peeters at the University of Montpellier, France; and Beatrice Hahn at the University of Pennsylvania.
The work was supported by funding from the National Institutes of Health, Agence Nationale de Recherche sur le Sida, Jane Goodall Institute, Arthur L. Greene Fund and Harvard University.
RELATED INFORMATION
