Berkeley-Led Study Identifies Human Lung Proteins That Can Advance or Thwart SARS-CoV-2 Infections
Researchers have taken an important step toward understanding the microscopic battle that plays out between our lung cells and the SARS-CoV-2 virus that causes COVID-19. A UC Berkeley-led study has identified specific proteins within our bodies that can promote or protect us from SARS-CoV-2 infections, potentially opening the door to new antiviral therapies.
In the study, published this week in Nature Genetics, researchers used CRISPR technology to test the impact of every human gene on SARS-CoV-2 infections in human lung cells. Their findings revealed new pathways that the virus relies on to infect cells, as well as the antiviral pathways that help protect against viral infection. Notably, they showed that mucins — the main component of mucus found in the lungs — seem to help block the SARS-CoV-2 virus from entering our cells.
“Our data suggest that mucins play a key role in restricting SARS-CoV-2 infection by acting as a barrier to viruses that are attempting to access our lung epithelial cells,” said Scott Biering, the study’s co-lead author and a postdoctoral researcher in Eva Harris’s lab at UC Berkeley’s School of Public Health. “Further, our data suggest that mucin expression levels in an individual’s lungs may impact COVID-19 disease progression.”
Patrick Hsu — cofounder of the Arc Institute and Berkeley assistant professor of bioengineering, Deb Faculty Fellow and Innovative Genomics Institute Investigator — is the principal investigator of the study, which brought together researchers from 10 institutions. Harris, professor of public health at UC Berkeley, and Silvana Konermann, assistant professor at Stanford University School of Medicine, were co-senior authors of the study. Other collaborators contributed from Stanford University, the University of North Carolina at Chapel Hill, Yale School of Medicine and Cornell University, and spanned disciplines ranging from immunology, bioengineering, epidemiology, molecular biology and genetics.
Together, the researchers were trying to determine how the SARS-CoV-2 virus enters human cells and replicates so efficiently during illness. They also wanted to identify specific defense mechanisms in human cells that might be able to fight infection, which could inspire new therapeutic strategies.
Researchers discovered that MUC1 and MUC4, types of mucins found in lung cell membranes, defend lung cells from infection. This finding is important because previous studies had suggested that an accumulation of mucus could be the reason why some people became seriously ill with COVID-19 — since the mucus can make it difficult for people to breathe — and proposed using drugs to deplete mucus. This Berkeley-led study suggests that such a strategy could interfere with mucins that provide a valuable defense mechanism against SARS-CoV-2 infection.
Still, mucins are complex, and more research is needed to fully understand them. The researchers discovered that other mucins — MUC5AC and MUC5B — which are secreted into the mucus lining of the lungs, either do nothing to stop SARS-CoV-2 infection or can even promote viral infection.
According to Biering, both the type and amount of mucus that each person produces may result in different outcomes for SARS-CoV-2 infection — and lead to different treatment strategies.
“Somebody who produces a lot of the right type of mucus could be very protected. But somebody who produces a lot of the wrong type of mucus might have more risk of infection,” said Biering. “And somebody who produces very little of the right type also could be at more risk.”
This is also the first study in which researchers looked at how the SARS-CoV-2 virus interacts with human lung cells, marking a critical advancement in COVID-19 research.
“It is well known that virus infection can be promoted or inhibited by our own proteins,” said Biering. “But this is the first time that a systematic investigation of these host cell proteins has been conducted in human lung epithelial cells for SARS-CoV-2 infection.”
Past studies of SARS-CoV-2 infection have used cell types that do not naturally contain the receptors or other pathways that the virus uses to infect our lungs. For this study, researchers wanted to use a more relevant cell type, human epithelial cells called Calu-3, found on the inner surface of the lung. Though this cell line is extremely challenging to work with, researchers managed to gain a more accurate understanding of the biology involved in human SARS-CoV-2 infections.
“[Using Calu-3 cells] was a huge advancement, given that these are very representative of the first cells the virus contacts and infects in humans, and it revealed new pathways not seen in other cell lines,” said co-lead author Sylvia Sarnik, an assistant specialist in Hsu’s lab at the time of the study. “Overall, this study is a step forward in understanding viral infection pathways and paves the way for research toward better treatments in the future.”
The researchers conducted genome-wide CRISPR gain-of-function and loss-of-function screening to eliminate or overexpress every gene in the human genome. Then they measured the impact of these gene expression changes on SARS-CoV-2 infection in human lung epithelial cells. Importantly, many of the genes highlighted by this screen have yet to be investigated experimentally, providing a starting point for future work.
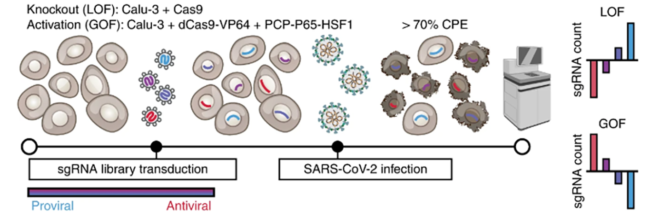
“You can run these massively parallel ‘Hunger Games’-style experiments on human cells to see which genes you can change to tune the ability of our lung cells to survive or grow when the virus infects them,” said Hsu.
“Our screen successfully identified hundreds of genes that were important for replication of SARS-CoV-2 and hundreds of genes that could restrict SARS-CoV-2,” said Biering. “In this study, we chose to focus on understanding the role of these mucin proteins.”
The researchers identified mucin glycoproteins as key restriction factors for infection in cells, both in mouse models and potentially in humans.
Researchers also studied how mucins would interact with other respiratory viruses, including influenza A virus, human parainfluenza virus, common cold coronaviruses and respiratory syncytial virus. As with SARS-CoV-2, the results were unpredictable.
“What we showed was mucins are broadly antiviral, but the story is actually much more complicated than that,” said Hsu. “In fact, in some viruses, we found that mucin overexpression actually seemed to increase infectivity.”
Future work will be critical to better understand how viruses interact with mucins, but, for now, these findings provide an important starting point. “This study helped us learn more about the virus and opened new avenues for research to further investigate druggable targets,” said Sarnik.
Fast Grants, the National Institutes of Health and the Centers for Disease Control and Prevention helped support this research.
