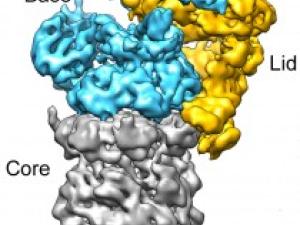
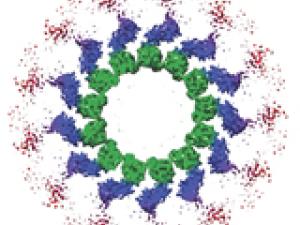
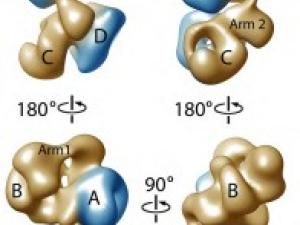Research Expertise and Interest
cryo-EM, biochemistry, complex biological assemblies, structure and regulation of the cytoskeleton, microtubule dynamics, human transcriptional initiation machinery, epigenetics, gene silencing, biophysics
Research Description
Eva Nogales is a Professor of Biochemistry, Biophysics and Structural Biology, and a Howard Hughes Investigator. The Nogales lab is dedicated to gaining mechanistic insight into crucial molecular processes in the life of the eukaryotic cell. Their two main research themes are the dynamic self-assembly of cytoskeleton during its essential functions in cell division, and the molecular machines governing the regulation of gene expression, specially at the transcriptional level. The unifying principle in their work is the emphasis on studying macromolecular assemblies as whole units of molecular function by direct visualization of their architecture, functional states and regulatory interactions. With this overall aim in mind they use electron microscopy and image analysis, complemented with biochemical and biophysical assays, towards a molecular understanding of their systems of interest.
Current Projects
Microtubule Structure and Dynamics
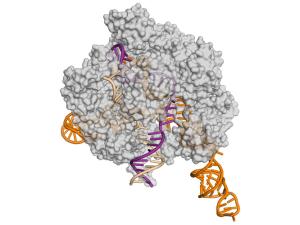
Microtubules are essential cytoskeletal polymers involved in processes as diverse as intracellular traffic and cell division. A long-standing interest of her lab concerns the process of microtubule dynamic instability and its role in mitosis. A large number of cellular factors regulate this dynamic behavior. We have defined the structure of the microtubule at the atomic level, and the conformational changes that accompany GTP hydrolysis and lead to microtubule destabilization, Their microtubule cytoskeleton studies also emphasize the dynamic interactions of microtubule with cellular factors, specially those involved in cell division. Many such factors, by interacting with the dynamic ends of microtubules, are able to modify the behavior of these polymers and to make use of the unique structures at microtubule ends to localize in space and time, or to change microtubule dynamics.
Regulation of Gene Expression: Transcription Initiation
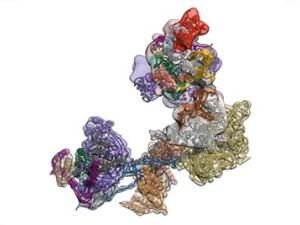
The regulated transcription of genes in all eukaryotes requires the assembly of a complex molecular machine around the transcription start site. Electron microscopy is ideally suited to the study large macromolecular complexes. The Nogales lab has used cryo-EM to define the structure and functional transitions of the human transcription preinitiation complex, which includes TFIID, TFIIA, TFIIB, TFIIE, TFIIF, TFIIH and RNA pol II. Their goal is now to characterize their mechanism of action in the context of chromatin, as well as their interaction activators, repressors and cofactors to generate an integrated, mechanistic model of transcriptional regulation.
Regulation of Gene Expression: Gene Silencing by PRC2
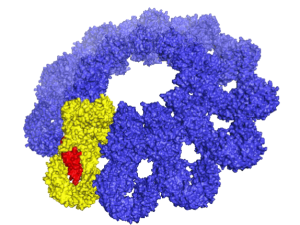
Polycomb Repressive Complex 2 (PRC2) is essential for gene silencing, establishing transcriptional repression of specific genes by tri-methylating Lysine 27 of histone H3, which ultimately lead to chromatin compaction. The NOgales lab have used cryo-EM to define the structure of PRC2 bound to two critical cofactors, and visualized the interaction of PRC2 bound to a dinucleosome. We are continuing our structural studies, complemented with activity assays, to explore the complex regulation that arises form the cross-talk between PRC2 dofferent cofactors and different chromatin state (post-translational modifications of histone tails).
In the News
Eva Nogales Wins Shaw Prize in Life Science and Medicine
AAAS Adds Ten New Fellows From UC Berkeley
Freeze-frame microscopy captures molecule’s ‘lock-and-load’ on DNA
New structure of tau protein, a key player in Alzheimer’s disease
Long-sought structure of telomerase paves way for drugs for aging, cancer
How the Immune System Identifies Invading Bacteria
Anti-CRISPR proteins decrease off-target side effects of CRISPR-Cas9
Scientists decipher opening dance steps of DNA and its partner
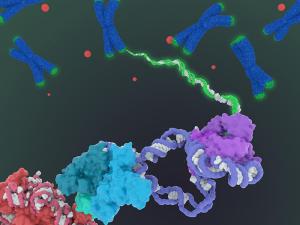
Working at temperatures near absolute zero, UC Berkeley experts in electron microscopy have learned in detail how proteins orchestrate the first key steps in gene activation – opening up the double-stranded DNA.
American Academy of Arts and Sciences elects nine Berkeley faculty members
The American Academy of Arts and Sciences today announced the election of 213 new members, including nine UC Berkeley faculty members.
Five Berkeley scientists named to National Academy
Discovery of how Taxol works could lead to better anticancer drugs
UC Berkeley scientists have discovered the extremely subtle effect that the prescription drug Taxol has inside cells that makes it one of the most widely used anticancer agents in the world.
New insight into an emerging genome-editing tool
Biochemist Jennifer Doudna and biophysicist Eva Nogales led an international collaboration with results that point the way to the rational design of new and improved versions of Cas9 enzymes for basic research and genetic engineering.
New Key to Organism Complexity Identified
The enormously diverse complexity seen amongst individual species within the animal kingdom evolved from a surprisingly small gene pool. The key to morphological and behavioral complexity, a growing body of scientific evidence suggests, is the regulation of gene expression by a family of DNA-binding proteins called “transcription factors.”
Berkeley Lab Scientists Create First 3-D Model of a Protein Critical to Embryo Development
The first detailed and complete picture of a protein complex that is tied to human birth defects as well as the progression of many forms of cancer has been obtained by an international team of researchers led by scientists with the Department of Energy, Berkeley Lab and UC Berkeley.
New Information on the Waste-Disposal Units of Living Cells
Berkeley researchers have provided the most detailed look ever at the “regulatory particle” used by the proteasome - one of the most critical protein machines in living cells - to identify and degrade proteins marked for destruction.
Focusing on a key event in life of a chromosome
If chromosomes are to separate properly during cell division, the microtubules in the cell must grab onto them and pull them apart. The place where the microtubules latch on is a large molecular complex called the kinetochore. Eva Nogales and her UC Berkeley/LBNL team have created the first high-resolution image of this junction and identified regulators that prevent errors that can lead to cancer or death.